Annals of Clinical Microbiology and Antimicrobials volume 24, Article number: 42 (2025) Cite this article
Sepsis remains a major public health challenge, leading to high mortality and morbidity rates, particularly among low-income populations such as those in sub-Saharan Africa. Its management is complicated by the emergence of multidrug-resistant bacterial strains, necessitating microbiological surveillance and adaptation of antibiotic therapy. This study examines the microbiological profile of sepsis and the antibiotic susceptibility of pathogens among critically ill patients in Lubumbashi, Democratic Republic of Congo.
A prospective study was conducted from January 2021 to December 2023 across three ICU units in Lubumbashi. Patients suspected of having sepsis were included, and microbiological samples were collected from various sources (blood, urine, pus, biological fluids). Bacterial identification and antibiotic susceptibility testing were performed according to Clinical and Laboratory Standards Institute (CLSI) guidelines. Data were analyzed using SPSS version 23® and Excel 365®.
Among the 76 patients included, 40% had confirmed bacterial sepsis. The predominant isolates were Gram-negative bacilli (62.7%), with Escherichia coli (28.35%) and Klebsiella pneumoniae (22.73%) being the most common species. Gram-positive bacteria accounted for 33.89%, primarily coagulase-negative streptococci (15.11%) and Enterococcus faecium (5.61%). Antimicrobial resistance profiles revealed a high level of resistance to commonly used antibiotics, particularly cephalosporins, fluoroquinolones, and cotrimoxazole. However, greater sensitivity was observed with amikacin (41.3%), fosfomycin (37%), and meropenem (33.8%).
This study highlights the high prevalence of Gram-negative bacteria and concerning resistance to first-line antibiotics, jeopardizing the effectiveness of empirical treatments. These findings underscore the urgency of strengthening microbiological surveillance, rationalizing antibiotic use, and implementing antimicrobial resistance control policies in the DRC. Developing treatment protocols tailored to local data and enforcing stricter antibiotic regulations are essential to improving sepsis management and reducing associated mortality.
Sepsis is a major global public health concern due to its high morbidity and mortality, as well as its economic impact [1, 2]. It causes more deaths than several major diseases combined, including prostate, breast, and cervical cancers, coronary artery disease, strokes, and HIV/AIDS [3, 4]. Sepsis is estimated to be responsible for approximately 20% of global deaths, with a particularly high burden in low-income countries, especially in sub-Saharan Africa [5]. Annually, around 17 million cases of sepsis and 3.5 million deaths are recorded in this region [6, 7]. In Africa, this infection primarily affects younger populations, leading to a substantial loss of life years [8].
Sepsis is characterized by organ dysfunction resulting from an exaggerated and dysregulated inflammatory response to infection, involving immune, endocrine, and metabolic disturbances [9,10,11]. It remains a critical issue, particularly in resource-limited settings, where it represents a significant cause of morbidity and mortality, especially in intensive care units [12]. Early diagnosis and prompt administration of appropriate antibiotics are crucial in reducing sepsis-associated mortality [13]. However, pathogen identification is challenging due to the variability in blood culture reliability, influenced by factors such as timing of sample collection and diagnostic techniques used [14].
Bacterial pathogens responsible for sepsis include Gram-positive species (Streptococcus pneumoniae, Staphylococcus aureus, Streptococcus agalactiae) and Gram-negative species (Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa) [15, 16]. Studies conducted in Africa, including Ethiopia, Uganda, and Egypt, report a predominance of Gram-negative bacteria with alarming antimicrobial resistance rates. In Ethiopia, Pseudomonas aeruginosa, Klebsiella pneumoniae, and Escherichia coli are commonly implicated, exhibiting resistance rates up to 80% [17, 18]. This scenario is exacerbated by unregulated antibiotic sales, self-medication, and excessive antimicrobial use in agriculture, all contributing to the spread of bacterial resistance [14].
Empirical antibiotic therapy remains the cornerstone of sepsis management. However, the growing prevalence of multidrug-resistant (MDR) strains challenges this approach [4]. In the DRC, several studies highlight inappropriate antibiotic use, driven by low awareness of antimicrobial resistance risks, frequent self-medication, and weak pharmaceutical regulatory frameworks. A study conducted in Kinshasa revealed that these practices are influenced by perceived infection risks, poverty, and inadequate drug control systems [19].
A study on 320 blood cultures in the DRC found that 92.9% of identified bacteria were Gram-negative, with Escherichia coli (51.9%), Klebsiella spp. (20.2%), and Enterobacter spp. (6.7%) being the most prevalent [20]. High resistance rates were observed against commonly used antibiotics, including cotrimoxazole (100%), erythromycin (100%), and ampicillin (66.7–100%), alongside increasing MDR strains and extended-spectrum beta-lactamase (ESBL)-producing Enterobacteriaceae.
Given these findings, strengthening microbiological surveillance and improving antibiotic regulation are crucial in the DRC. Previous studies emphasize the urgent need for national antimicrobial resistance control strategies, including the establishment of laboratories and surveillance networks to mitigate sepsis-related mortality [21]. Furthermore, a study in Eastern DRC reported severe resource shortages for sepsis management, including limited oxygen supply, broad-spectrum antibiotics, and diagnostic tools [22]. These structural challenges hinder the implementation of international guidelines, such as the Surviving Sepsis Campaign (SSC) recommendation [23], necessitating urgent interventions.
This study was conducted within this context. Its objective is to assess the bacteriological profile of sepsis and antimicrobial susceptibility patterns among ICU patients in Lubumbashi, DRC. Additionally, it aims to understand the factors contributing to antimicrobial resistance and guide strategies for effective sepsis management in this public health challenge.
This was a prospective, multicenter descriptive study conducted over a three-year from January 2021 to December 2023 in the multipurpose intensive care units of three hospitals in Lubumbashi, Democratic Republic of Congo: the Cliniques Universitaires de Lubumbashi (CUL), the SNCC Hospital (Hopital de la Société Nationale des Chemins de Fer du Congo), and the Centre Médical Diamand de Lubumbashi(CMDL). These units were selected based on the presence of an anesthetist-intensive care unit, high patient admission rates, their role as referral centers, and the availability of basic intensive care equipment. The study group consisted of patients admitted for more than 48 h to the medical and surgical intensive care units.
The study included all patients admitted to the ICU or Continuing Care Unit (CCU) with suspected sepsis during the study period. These patients required intensive monitoring and treatment due to their critical condition.
Sampling was conducted on a random and non-exhaustive basis, determined by available resources, including staffing and funding constraints. The estimated number of admissions and sepsis cases was based on hospital data, identifying 76 cases over the study period.
Various clinical specimens were collected to identify potential pathogens. Table 1 summarizes Types of biological samples collected from patients based on suspected infection sources. The table describes sample counts, collection methods, and clinical indications guiding microbiological investigations.
Blood samples (2 to 5 mL) were inoculated into aerobic culture vials (BACT/ALERT®, BioMérieux, France) and incubated at 37 °C for up to 7 days. Sample processing, pathogen identification to the species level, and antimicrobial susceptibility testing were performed according to standard laboratory protocols. Initial samples underwent Gram staining and were cultured on blood agar and MacConkey agar, incubated aerobically at 37 °C for 18 to 24 h. Bacterial isolates were identified based on colony morphology and biochemical characteristics.
In addition to blood cultures, all other clinical specimens, including urine, pus, bronchial aspirates, cerebrospinal fluid (CSF), catheter tips, and pleural or ascitic fluids, were processed following standard microbiological protocols. Urine samples were inoculated onto CLED and MacConkey agars and incubated aerobically at 37 °C for 18 to 24 h. Pus samples and wound swabs were cultured on blood and MacConkey agars under the same incubation conditions. Bronchial aspirates and CSF specimens were directly inoculated on blood agar, chocolate agar, and MacConkey agar, depending on the suspected pathogens. Central venous catheter tips were cultured on blood agar using the semi-quantitative Maki roll-plate method. Pleural and ascitic fluids were centrifuged, and the resulting sediment was inoculated on blood and MacConkey agars. All plates were incubated aerobically at 37 °C and examined after 24 and 48 h for colony growth. Identification of bacterial isolates was based on colony morphology, Gram staining, and standard biochemical tests.
Fungal pathogens were evaluated through direct microscopic examination (wet mount and Gram staining) and cultured on Sabouraud Dextrose Agar, incubated at 28–30 °C for up to 7 days. Fungal identification was based on colony appearance and microscopic characteristics. However, no systematic evaluation for viral, malarial, or Mycobacterium tuberculosis infections was performed due to limited access to advanced diagnostic tools such as PCR, rapid antigen tests, or Ziehl-Neelsen staining.
Antimicrobial susceptibility testing was performed on Mueller-Hinton agar using Kirby-Bauer’s disk diffusion method. Commercial antibiotic discs (HiMedia Laboratories, Mumbai, Maharashtra, India) were used, including ampicillin, erythromycin, azithromycin, cefotaxime, ceftriaxone, cefepime, gentamicin, amikacin, ciprofloxacin, levofloxacin, ofloxacin, piperacillin/tazobactam, imipenem, meropenem, co-trimoxazole, tetracycline, vancomycin, nitrofurantoin, and fosfomycin. Inhibition zones were measured after 24 h of incubation at 35–37 °C and interpreted according to CLSI thresholds.
Sepsis was defined according to the Sepsis-3 criteria as life-threatening organ dysfunction caused by a dysregulated host response to infection. Organ dysfunction was identified by an increase in the Sequential Organ Failure Assessment (SOFA) score of ≥ 2 points from baseline [24]. In clinical settings where full SOFA calculation was not immediately available, the quick SOFA (qSOFA) score, based on systolic blood pressure ≤ 100 mmHg, respiratory rate ≥ 22/min, and altered mental status (Glasgow Coma Scale < 15), was used for initial bedside screening. Confirmation of sepsis required documentation of infection and full SOFA score assessment.
Data were entered into Microsoft Excel and statistically analyzed using SPSS version 23. Descriptive statistics were calculated as frequencies for categorical variables.
The study protocol was approved by the Ethics Committee of the University of Lubumbashi under reference number UNILU/CEM/030/2021. Approval letters were also obtained from hospital directors. Voluntary informed consent was obtained from each patient before participation, following the Helsinki Declaration guidelines. Confidentiality and anonymity were strictly maintained during data collection and processing by the research team.
Table 2. Baseline Characteristics of ICU Patients with Suspected Sepsis (n = 76).
Demographic and clinical characteristics of the 76 patients admitted to intensive care units with suspected sepsis. Variables include age, sex, comorbidities, duration of ICU stay, need for mechanical ventilation, and in-hospital mortality.
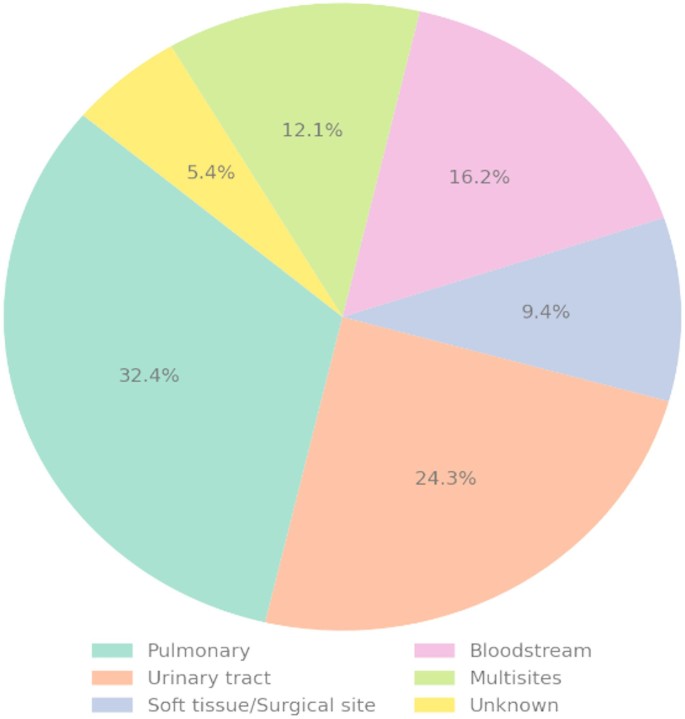
The most frequently identified suspected sources of infection were pulmonary infections (28.6%) and urinary tract infections (26.0%), followed by multisite infections (18.0%) and bloodstream infections (11.7%). Surgical site or soft tissue infections accounted for 8.4% of cases. Less commonly reported sources included abdominal infections (2.3%), other origins (2.0%), and unidentified sources (3.0%). This distribution underscores the predominance of respiratory and urogenital infections in the pathogenesis of sepsis among ICU patients (Fig. 1).
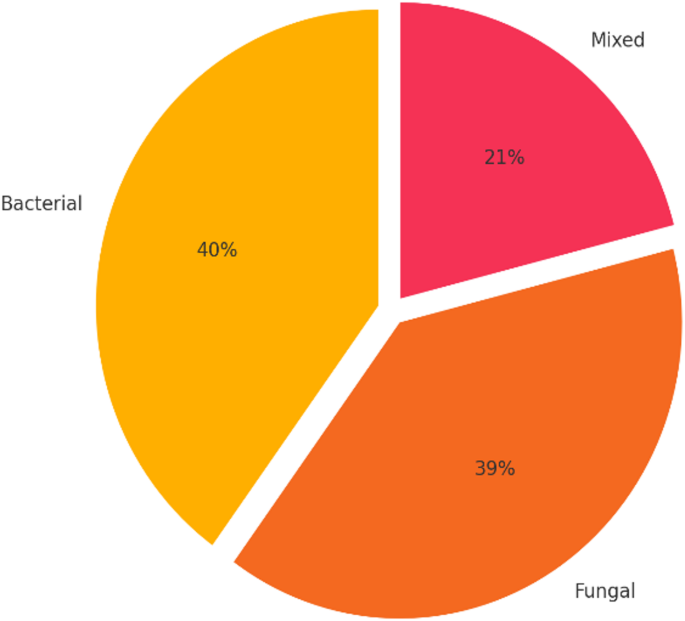
Microbiological cultures were performed on blood, sputum, and various body fluids (including urine and pus) obtained from 76 patients with suspected sepsis. Each specimen was collected based on the clinically suspected site of infection and guided the microbiological investigation accordingly. Figure 2. illustrates the distribution of infections by type, distinguishing bacterial, fungal, and mixed infections. Of the 76 sepsis cases analyzed, 30 (40%) involved pure bacterial infections, 29 (39%) were due to fungal pathogens, and 17 cases (21%) involved mixed infections with both bacterial and fungal agents. These were identified using direct microscopic examination (wet mount and Gram staining) and cultured on Sabouraud Dextrose Agar incubated at 28–30 °C for 5–7 days. Colony morphology and microscopic characteristics were used for species identification. These findings highlight the significant contribution of fungal and co-infections in the overall infectious burden among critically ill patients.
Table 3 presents the distribution of microorganisms identified in this study. Gram-negative bacteria (GNB) accounted for 69.93% of isolates, with Escherichia coli (28.35%) and Klebsiella pneumoniae (22.73%) being the most common pathogens. These findings align with global trends, where GNB are frequently implicated in nosocomial and community-acquired infections. Gram-positive bacteria (GPB) represented 30.07% of isolates, primarily Coagulase-Negative Staphylococcus (15.11%), followed by Enterococcus faecium and Enterococcus faecalis and Staphylococcus aureus (1.87%). The presence of Pseudomonas aeruginosa (7.62%) and Citrobacter spp. (5.62%) highlights the diversity of the pathogens involved.
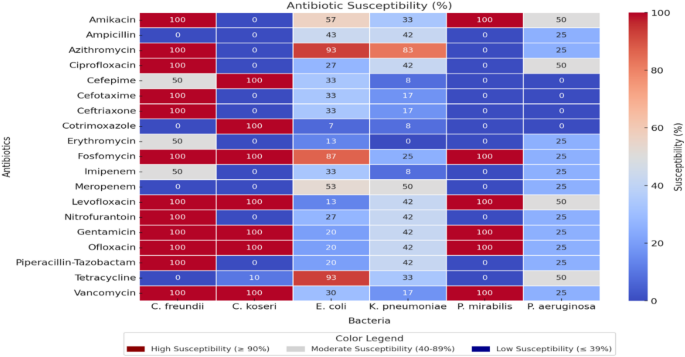
The identified pathogens were tested for their susceptibility to specific antimicrobials, and Fig. 3 presents the susceptibility rates (%) of each bacterium to a range of antibiotics in the form of a heat map. Overall, Amikacin and Fosfomycin exhibit high susceptibility rates (≥ 90%) against C. freundii and P. mirabilis, whereas Ampicillin and Meropenem demonstrate low efficacy (≤ 39%) against certain strains (C. koseri and C. freundii). Fluoroquinolones (e.g., Ciprofloxacin, Ofloxacin) and certain cephalosporins (e.g., Cefepime, Cefotaxime) display moderate susceptibility rates (40–89%) against E. coli and K. pneumoniae but are weakly effective (≤ 39%) against P. aeruginosa. Additionally, Azithromycin and Levofloxacin maintain high susceptibility (≥ 90%) against certain Enterobacteriaceae, while antibiotics such as Gentamicin and Ciprofloxacin exhibit partial efficacy (40–89%). Finally, the observed complete inefficacy (≤ 39%) of Ampicillin, Imipenem, or Cefepime in certain cases underscores the importance of tailoring treatment protocols based on local susceptibility profiles to ensure optimal infection management.
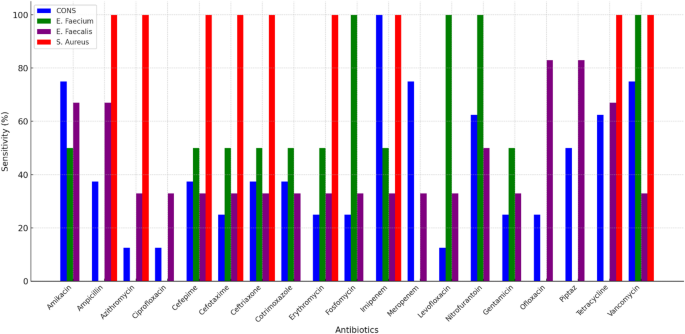
Figure 4 illustrates the susceptibility of major Gram-positive bacteria to the tested antibiotics, highlighting significant interspecies variations. Staphylococcus aureus demonstrated high susceptibility (> 95%) to Vancomycin, Imipenem, and Fosfomycin, confirming their efficacy against S. aureus infections, including methicillin-resistant strains (MRSA). In contrast, Enterococcus faecium exhibited alarming resistance, with susceptibility rates below 30% for beta-lactams, fluoroquinolones, and macrolides, indicating limited therapeutic options.
Although Enterococcus faecalis was more susceptible than E. faecium, it showed high resistance (> 60%) to cephalosporins and fluoroquinolones, excluding them as first-line treatments. However, its susceptibility to Vancomycin remained high (> 90%), making it a preferred option for severe infections. Coagulase-negative staphylococci (CONS) displayed intermediate susceptibility, with notable effectiveness of Vancomycin (85%) and Tetracycline (70%), despite the presence of resistance exceeding 50% for ampicillin and fluoroquinolones.
The objective of this study was to determine the bacterial profile associated with sepsis and to assess the antimicrobial resistance patterns among ICU patients in Lubumbashi, Democratic Republic of the Congo (DRC). Our findings reveal several clinically significant insights that have both local and regional implications for patient management and public health strategy.
Sepsis remains a major challenge in critical care, particularly in resource-limited settings where diagnostic infrastructure and antimicrobial stewardship practices are often lacking. Microbiological analysis of samples from 76 patients revealed a polymicrobial profile, with bacterial isolates accounting for 40% of positive cases. This confirms their dominant role in systemic infections and is consistent with previous literature [25]. Notably, fungal pathogens were isolated in 39% of cases, significantly higher than the 15–16% typically reported globally [26,27,28].
These findings suggest that fungal sepsis may be significantly underrecognized in our setting, likely due to gaps in empirical treatment strategies and limited access to appropriate diagnostics. In many low-resource environments, fungal infections in sepsis are often overlooked. Most African studies, such as those by Yimtchi et al. and Abera et al., primarily focus on bacterial pathogens and seldom incorporate fungal diagnostic [13, 14]. This omission likely contributes to the underreporting of fungal sepsis in sub-Saharan Africa, where access to fungal culture media and biomarkers remain scarce.
In contrast, several studies in Asia have reported a rising incidence of fungal infections in ICU sepsis. For instance, Sharma et al. documented bacterial infections in 57% of cases, fungal infections in 27%, and mixed infections in 16% in an Indian ICU [29]. Similarly, Bhabhor et al. reported that fungal bloodstream infections affected approximately 30% of ICU patients, with Candida albicans as the predominant species [30]. Although the prevalence reported in these studies is lower than in our cohort, they underscore the increasing global importance of fungal pathogens in critical care.
Fungal sepsis is well known to be associated with poorer outcomes, such as prolonged ICU stays, higher mortality, and elevated healthcare costs. These outcomes are often driven by diagnostic delays and the failure of empirical antibacterial therapies to target fungal agents [31]. Therefore, integrating fungal screening into routine sepsis protocols, particularly for patients who fail to respond to antibiotics, is essential. Our results highlight the urgent need to broaden sepsis diagnostics to include fungal pathogens and to consider early antifungal therapy in high-risk patients. Such an approach is vital to improving outcomes and reducing sepsis-related mortality, especially in resource-constrained settings.
The prevalence of bacterial infections observed in this study aligns with data reported by Chakraborty et al. in India and Nasrin et al. in Bangladesh, who identified bacteria as the predominant pathogens in both hospital and community-acquired infections. Escherichia coli and Klebsiella pneumoniae are frequently implicated, particularly in urinary tract, respiratory, and bloodstream infections [32, 33]. Similar trends have been reported in Egypt (32.37%) and Ethiopia (25.78%), highlighting the global significance of this issue [12, 34].
The findings of the present study confirm the predominance of Gram-negative bacteria (GNB) among the pathogens responsible for bacterial infections and sepsis, accounting for 69.93% of the isolates. Escherichia coli (28.35%) and Klebsiella pneumoniae (22.73%) were the most frequently identified pathogens, a trend consistent with observations in other geographic regions, including Japan, Pakistan, and Egypt, where these bacteria are commonly isolated in sepsis cases [35, 36]. Similarly, a study conducted in Tanzania reported a high prevalence of E. coli and Pseudomonas aeruginosa in severe infections, underscoring the similarities in epidemiological patterns across different regions. The presence of Pseudomonas aeruginosa (7.62%) and Citrobacter spp. (5.62%) highlights the diversity of pathogens involved in both nosocomial and community-acquired infections [37]. P. aeruginosa is particularly concerning due to its multidrug resistance, including resistance to beta-lactams and fluoroquinolones, as reported by Tängdén et al. and Maina et al. [38, 39]. Additionally, Citrobacter spp. is an emerging opportunistic pathogen, particularly in immunocompromised patients, corroborating the observations of Ouedraogo et al. in Burkina Faso [40].
Gram-positive bacteria (GPB) accounted for 30.07% of the isolates, with coagulase-negative staphylococci (CONS) being the most prevalent (15.11%), followed by enterococci (Enterococcus faecium and E. faecalis) and Staphylococcus aureus (1.87%). This distribution aligns with findings from Chakraborty et al. in India and Rehman et al. in Bangladesh, who reported an increasing prevalence of coagulase-negative staphylococci (CONS) and enterococci in hospital settings, particularly in intensive care units. These pathogens are frequently associated with the use of vascular catheters and invasive medical devices, highlighting their role in healthcare-associated infections [32, 36].
The findings on pathogen susceptibility to antibiotics confirm trends reported in recent literature regarding the increasing resistance of Gram-negative bacteria. The high efficacy of Amikacin and Fosfomycin (> 90%) against Citrobacter freundii and Proteus mirabilis aligns with studies by Nasrin et al. and Gautam et al., which highlighted their sustained effectiveness against these urinary and gastrointestinal pathogens [33, 41]. This high susceptibility suggests that these antibiotics remain valuable options for the treatment of beta-lactam-resistant Enterobacteriaceae infections. Regarding Levofloxacin and Azithromycin, their strong activity against Escherichia coli and Klebsiella pneumoniae corroborates the findings of Ouedraogo et al. in Burkina Faso and Huynh et al., who recommend their use for the treatment of urinary tract and respiratory infections caused by these pathogens [40, 42]. However, the alarming resistance of E. coli and K. pneumoniae to Ciprofloxacin (73% and 42%, respectively) underscores the rapid increase in fluoroquinolone resistance, a phenomenon previously reported by Rehman et al. in Bangladesh and Chelkeba et al. in Ethiopia [36, 43]. This trend emphasizes the urgent need to limit the empirical use of fluoroquinolones and prioritize susceptibility testing before prescription to ensure effective treatment strategies.
The study highlights a concerning resistance to third- and fourth-generation cephalosporins in Escherichia coli and Klebsiella pneumoniae, with resistance rates reaching 92% for Cefepime and 83% for Cefotaxime. These findings align with those of Chakraborty et al., who reported a rising prevalence of extended-spectrum β-lactamase (ESBL)-producing Enterobacteriaceae in hospital settings, significantly compromising the efficacy of cephalosporins and limiting treatment options [32]. As a result, the increased use of carbapenems has become necessary for treating severe infections. However, their overuse contributes to the accelerated emergence of carbapenemase-producing strains, a phenomenon already observed by Vlieghe et al. in Central Africa. This underscores the urgent need for antimicrobial stewardship programs to optimize antibiotic use and curb the spread of multidrug-resistant pathogens [44].
The situation is even more critical for Pseudomonas aeruginosa, which exhibits pronounced multidrug resistance, with 100% resistance to Cefotaxime and Cefepime. This alarming trend, extensively documented in studies by Maina et al. and Tängdén, underscores the urgent need for rigorous microbiological surveillance and strict regulation of cephalosporin use against P. aeruginosa [38, 39]. Furthermore, while certain antibiotics such as Ciprofloxacin and Gentamicin retain partial efficacy (40–80% susceptibility), their markedly low effectiveness against E. coli (only 20%) suggests that they should not be used as monotherapy for severe infections. This aligns with the recommendations of Ikuta et al., who advocate for combination therapy and antimicrobial stewardship to mitigate the risks of treatment failure and further resistance development [45].
The finding that 86.7% of bacterial isolates exhibited multidrug resistance (MDR) underscores the critical scope of this issue. Similar resistance patterns to first-line antibiotics, such as Ampicillin, Azithromycin, and Cephalosporins, have been widely reported across Africa and Asia, indicating a broader regional trend and poses a significant challenge to the effective management of sepsis. These findings emphasize the urgent need for strengthened antimicrobial stewardship, regular surveillance of resistance patterns, and the development of alternative treatment strategies to combat the growing threat of MDR pathogens [34, 36, 46].
Additionally, the production of extended-spectrum β-lactamases (ESBL) in 64.6% of Enterobacteriaceae reflects a global trend, particularly in resource-limited regions, where the management of bacterial infections is increasingly challenged by resistance to conventional treatments [46, 47]. Furthermore, the complete failure of Meropenem and Ampicillin against certain strains, Notably, certain strains such as Citrobacter freundii and Citrobacter koseri demonstrated 100% resistance to both meropenem and ampicillin, indicating a complete lack of susceptibility. These findings highlight the presence of bacterial populations with extensive drug resistance, likely due to both intrinsic mechanisms and acquired resistance genes. The total non-susceptibility to these key antibiotics underscores the urgent need for routine susceptibility testing and the implementation of targeted antimicrobial stewardship interventions. This finding aligns with reports by Dagnew et al. in Ethiopia and Moore et al. in Uganda, further emphasizing the urgent need for targeted antimicrobial strategies and routine susceptibility testing to guide effective treatment decisions [48, 49].
The analysis of Gram-positive bacteria (GPB) in this study highlights the high susceptibility of Staphylococcus aureus to vancomycin (> 95%), contrasted with its pronounced resistance to β-lactam antibiotics. This resistance pattern is indicative of the widespread prevalence of methicillin-resistant Staphylococcus aureus (MRSA) in ICU settings. Similar trends have been documented in multiple studies across low- and middle-income countries. For instance, Jain et al., reported vancomycin susceptibility above 90% in MRSA isolates from ICU patients in India, reinforcing its continued role as a cornerstone of MRSA therapy [50]. Likewise, Nasrin et al., in Bangladesh found a high prevalence of MRSA among bloodstream isolates, with limited treatment options beyond glycopeptides such as vancomycin and teicoplanin [33]. These findings are consistent with our observations and underscore the urgent need for routine MRSA screening, particularly in ICU environments where invasive procedures and medical devices increase the risk of colonization and infection. Furthermore, the continued effectiveness of vancomycin must be safeguarded through judicious prescribing practices and active antimicrobial stewardship, especially in resource-limited settings where therapeutic alternatives are scarce.
Regarding enterococci, Enterococcus faecium exhibits significant resistance (> 70%) to cephalosporins and fluoroquinolones, consistent with the findings of Habyarimana et al. and Boussina et al., who reported increasing challenges in treating these infections due to the emergence of resistance to carbapenems and glycopeptides [51, 52]. However, Enterococcus faecalis retains a high susceptibility to Vancomycin (> 90%), aligning with the observations of Doare et al., who recommend Vancomycin as a therapeutic alternative for severe multidrug-resistant enterococcal infections. These findings highlight the critical need for antimicrobial stewardship and targeted therapy to preserve the efficacy of last-line antibiotics against resistant enterococcal strains [53].
Coagulase-negative staphylococci (CONS), frequently implicated in nosocomial infections, exhibit intermediate resistance to fluoroquinolones and Ampicillin (> 50%), a trend corroborated by studies conducted by Chelkeba et al. and Maina et al., who reported a rising prevalence of multidrug resistance in these hospital-associated pathogens [39, 43]. These findings underscore the critical need for continuous surveillance of resistance patterns, particularly given the increasing emergence of multidrug-resistant bacteria. Strengthening infection control measures and implementing antimicrobial stewardship programs are essential to mitigate the spread of resistant coagulase-negative staphylococci(CONS) and ensure effective treatment strategies in healthcare settings.
Another concerning trend observed in this study is the high resistance of Staphylococcus aureus and coagulase-negative staphylococci (CONS) to multiple antibiotics, although Vancomycin remains effective against these resistant strains. This pattern has also been reported in studies conducted in India and Ethiopia, where Vancomycin continues to be the first-line treatment for these pathogens [30, 54]. However, the excessive use of Vancomycin poses a significant risk of selecting for Vancomycin-intermediate (VISA) and Vancomycin-resistant S. aureus (VRSA) strains, necessitating careful antimicrobial stewardship in hospital settings. These findings highlight the need for judicious Vancomycin use, routine susceptibility testing, and the exploration of alternative therapeutic options to mitigate the risk of resistance development.
The antimicrobial resistance patterns identified in this study are consistent with a growing body of evidence from low- and middle-income countries, particularly in sub-Saharan Africa and Asia, where similar trends of multidrug resistance among Gram-negative and Gram-positive pathogens have been reported. For instance, Chakraborty et al. in India documented high levels of resistance to third-generation cephalosporins and fluoroquinolones among E. coli and K. pneumoniae isolated from ICU patients, mirroring our findings of 83–92% resistance to cefotaxime and cefepime [32]. Similarly, Rehman et al. in Bangladesh and Chelkeba et al. in Ethiopia observed alarming resistance rates to ciprofloxacin in E. coli and K. pneumoniae (up to 70%), closely matching the 73% and 42% resistance rates identified in our isolates [36, 43]. In East Africa, Maina et al. reported extensive β-lactam and carbapenem resistance in Pseudomonas aeruginosa, a pattern echoed in our study where P. aeruginosa showed 100% resistance to cefepime and cefotaxime [39]. Ouedraogo et al. in Burkina Faso also found E. coli and K. pneumoniae strains with high levels of ESBL production and partial susceptibility to aminoglycosides, comparable to our observed ESBL rate of 64.6% and preserved activity of amikacin [40].
A recent systematic review by Kiya et al., revealed a high prevalence of sepsis among hospitalized adults in sub-Saharan Africa, with a significant associated mortality rate. The study emphasized the diversity of pathogens and the rising trend of antimicrobial resistance (AMR) in the region, thereby corroborating the patterns observed in our research [55]. A scoping review on ICU infections in low- and middle-income countries (LMICs) further confirmed high rates of nosocomial infections and AMR, particularly involving multidrug-resistant species such as Acinetobacter baumannii, Pseudomonas aeruginosa, and Klebsiella pneumoniae [56].
In the Democratic Republic of the Congo, a study conducted at Bukavu General Hospital reported a predominance of Gram-negative bacteria (92.9%) in bloodstream infections, with Escherichia coli (51.9%) and Klebsiella spp. (20.2%) being the most frequently isolated. Alarmingly high resistance levels were found against co-trimoxazole (100%), erythromycin (100%), and ampicillin (66.7–100%), with moderate to high resistance to ciprofloxacin, ceftazidime, ceftriaxone, cefuroxime, and cefepime. A significant proportion of isolates exhibited multidrug resistance and ESBL production phenotypes [20].
When interpreting antimicrobial susceptibility patterns, it is crucial to differentiate between intrinsic and acquired resistance. Intrinsic resistance refers to the innate capacity of certain bacterial species to withstand specific antibiotics due to inherent structural or physiological characteristics. Conversely, acquired resistance arises from genetic mutations or horizontal gene transfer, enabling bacteria to resist antibiotics that were previously effective [57]. A notable example is Enterococcus faecium, which is intrinsically resistant to third-generation cephalosporins owing to the absence of appropriate penicillin-binding proteins [58]. Similarly, Pseudomonas aeruginosa displays innate resistance via the activity of efflux pumps and low outer membrane permeability [59, 60]. These mechanisms, while not always detectable through standard antimicrobial susceptibility testing, must be considered in result interpretation, especially in resource-limited settings where advanced diagnostic technologies are unavailable. Accordingly, the high resistance rates to cephalosporins observed in these species in our study should be interpreted as a reflection of well-established intrinsic resistance rather than as an indication of emerging or locally acquired resistance. Recognizing this distinction is essential for guiding empirical therapy and for effective antimicrobial resistance (AMR) surveillance.
Considering these findings, it is imperative to consider how AMR influences sepsis outcomes in the Democratic Republic of the Congo (DRC). The increasing prevalence of resistant bacterial strains in intensive care units across the DRC highlights several systemic challenges, including widespread empirical antibiotic use in the absence of microbiological confirmation, limited implementation of antimicrobial stewardship initiatives, inadequate infection prevention and control measures, and a lack of reliable diagnostic capacity. In many sub-Saharan African settings, including the DRC, inappropriate antibiotic use and weak resistance surveillance systems significantly contribute to this alarming trend [21, 61].
AMR poses a substantial threat to effective sepsis management. Delays in initiating appropriate antimicrobial therapy—whether due to resistant pathogens or unavailable culture results, are strongly associated with increased mortality. In the present study, the elevated mortality observed among septic patients may be partly attributed to such therapeutic delays and the inability to promptly adapt treatments based on resistance profiles.
To combat this critical issue, the DRC must prioritize the development of robust laboratory infrastructure, ensure routine microbiological testing and susceptibility profiling in ICUs, and establish a functional national AMR surveillance system. Enhancing infection control strategies and promoting rational antibiotic use through stewardship programs are also key interventions. As emphasized by the World Health Organization (2014), both global and national coordination are essential to mitigate the devastating consequences of AMR, particularly in low-resource countries like the DRC.
Our findings further highlight the indispensable role of continuous microbiological surveillance in healthcare settings. Studies conducted in ICUs of Zagazig University Hospital (Egypt) and Mangaluru Hospital (Pakistan) have shown that routine monitoring of nosocomial infections enables better alignment of empirical treatments with local resistance trends [46, 62]. In this context, rapid pathogen identification and timely adjustment of antimicrobial regimens based on culture and susceptibility data are critical for improving clinical outcomes. Strengthening microbiological surveillance and integrating real-time resistance data into clinical decision-making are therefore foundational steps toward enhancing infection control and antibiotic stewardship in hospital environments.
The rapid escalation of antimicrobial resistance (AMR) in the Democratic Republic of Congo (DRC), particularly in urban centers such as Lubumbashi, constitutes a pressing public health emergency. This growing threat is directly associated with increasing mortality rates from sepsis and other life-threatening infections. The persistence and amplification of AMR in the DRC are driven by a multifaceted combination of systemic healthcare deficiencies, microbiological trends, community-level behaviors, and broader contextual challenges. Together, these factors significantly compromise the effectiveness of infection prevention strategies and undermine antimicrobial stewardship interventions.
In the specific context of the DRC, several underlying drivers contribute to both the emergence and sustained transmission of AMR. A study conducted in Kinshasa by Pungu Shembo et al., highlighted the widespread inappropriate use of antibiotics at the community level. This misuse is fueled by limited public awareness of proper antibiotic indications, a high perceived risk of infection, and pervasive self-medication practices. Structural constraints, including poverty and the lack of effective regulatory mechanisms to ensure pharmaceutical quality—further exacerbate irrational antibiotic use. These behaviors promote the selection and spread of resistant pathogens across communities [19].
In recognition of these challenges, a national call to action has emphasized the urgent need to implement robust AMR control strategies in the DRC. Priority interventions include the development of a nationwide surveillance infrastructure, investment in laboratory capacity, and the promotion of coordinated multisectoral approaches involving human health, veterinary, and environmental sectors. Surveillance remains a cornerstone in this response, enabling real-time detection of resistant infections, informing evidence-based treatment protocols, and guiding national public health policy [21].
This study has several limitations that may affect the generalizability and interpretation of the results. First, it was conducted in three hospitals in Lubumbashi, limiting the representativeness of the findings to this specific region and tertiary care settings, potentially excluding patients from rural areas or those without access to these facilities. Additionally, the sampling was random but not exhaustive, which may introduce selection bias. The study duration was restricted to three years from January 2021 to December 2023, preventing the assessment of seasonal variations or potential changes in hospital practices and antibiotic usage over time. Furthermore, although strict inclusion and exclusion criteria were applied, the data collection methodology may have introduced biases. Specifically, reliance on culture-based pathogen identification could underestimate the presence of certain pathogens, while the absence of molecular techniques limited the detection of specific resistance mechanisms.
Moreover, this study did not investigate viral, malarial, or tuberculosis-related etiologies, which may also contribute to sepsis, particularly in endemic regions such as the Democratic Republic of Congo. The lack of specific diagnostic tools, such as PCR, rapid antigen tests, or molecular assays, limited our ability to detect these non-bacterial pathogens and represents a key limitation of the present study. In addition, the study did not explore risk factors associated with antimicrobial resistance, including comorbidities, prior antibiotic exposure, or length of ICU stay, all of which could have provided valuable insights into resistance patterns. Despite these limitations, the findings underscore the urgent need to strengthen microbiological surveillance, improve diagnostic capabilities, and promote the rational use of antimicrobials to combat the escalating threat of antimicrobial resistance in low-resource settings.
This study provides crucial insights into the complex and multifactorial nature of sepsis in intensive care units in Lubumbashi, Democratic Republic of the Congo. The significant burden of both bacterial and fungal infections, including a substantial proportion of mixed infections—highlights the need for expanded diagnostic approaches and adapted therapeutic strategies. In particular, the predominance of certain fungal pathogens underlines the importance of integrating fungal screening into routine sepsis management, especially for critically ill and immunocompromised patients.
The high prevalence of multidrug-resistant Gram-negative bacteria poses a serious challenge to effective treatment and underscores the urgency of antimicrobial resistance (AMR) containment. The distinction between intrinsic and acquired resistance is critical to avoid misinterpretation of susceptibility data and inappropriate empirical therapy. In the DRC context, structural limitations such as insufficient laboratory capacity, lack of routine surveillance, and absence of antimicrobial stewardship programs contribute significantly to poor outcomes and high mortality rates.
Addressing these challenges requires a coordinated and comprehensive response. This includes strengthening microbiological diagnostics, implementing national AMR surveillance, promoting rational antibiotic use, and enforcing infection prevention and control (IPC) measures. Policy-level actions are also needed, such as stricter regulation of antibiotic distribution, improved healthcare provider training, and public awareness initiatives.
Ultimately, improving the management of sepsis and controlling antimicrobial resistance in the DRC will depend on sustained investments in health system strengthening, the integration of local epidemiological data into clinical practice, and the implementation of evidence-based guidelines. These efforts are essential to preserve the effectiveness of available treatments, ensure better patient outcomes, and reduce sepsis-related mortality in resource-limited settings.
No datasets were generated or analysed during the current study.
The authors are grateful for the assistance provided by the medical and nursing teams of the intensive care units of the three aforementioned hospitals.
This study did not receive any specific funding from commercial or non-commercial sources. The authors covered all costs associated with this research.
The authors declare no competing interests.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
Manika, M.M., Tano, A.K., Kapend’a, L.K. et al. Bacteriological profile and antimicrobial resistance in sepsis cases in intensive care units in Lubumbashi: challenges and perspectives. Ann Clin Microbiol Antimicrob 24, 42 (2025). https://doi.org/10.1186/s12941-025-00811-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12941-025-00811-w








-1752733979579.webp)


