BMC Veterinary Research volume 21, Article number: 351 (2025) Cite this article
Antimicrobial resistance (AMR) is a major global challenge that disproportionately affects low- and middle-income countries (LMICs). Environmental contamination by resistant bacteria from animal production facilities is a major driver of the spread of AMR through the food chain, requiring a robust one-health control approach. Traditional culture-based AMR surveillance is time-consuming and less sensitive, and fails to fully capture the spectrum of AMR, evolutionary trends, and epidemiological patterns of AMR spread. Whole-genome sequencing (WGS) has revolutionized AMR surveillance capabilities. Rapid WGS captures the full AMR spectrum with minimum samples, aids source attribution, and provides insights into trends in AMR spread. The portable Oxford Nanopore® Technology (ONT) platform, coupled with open-source software such as Galaxy and Konstanz Information Miner (KNIME), enables the establishment of a potentially portable, transferable workflow for low-resource settings. This study aimed to assess the AMR burden on four dairy farms in Kandy, Sri Lanka, via a resource-limited LMIC using a low-cost high-throughput screening assay and rapid WGS via ONT with Galaxy and KNIME processing to obtain full antibiotic resistomes.
The four isolates exhibiting the highest minimum inhibitory concentrations for amoxicillin were identified as Enterobacter cloacae and E. hormaechei by WGS. Chromosomes (4.8 to 4.9 Mb) carry the strain-specific resistance genes blaCMH-1, blaACT-25, fosA_7, and ramA, which are associated with diverse antibiotic classes. Plasmids, including IncFIB (pECLA), IncFII (pECLA), and IncX3, carry multiple resistance genes, including AAC(3)-IIe, AAC(6’)-Ib-cr, APH(3″)-Ib, APH(6)-Id, blaCTX-M-15, blaNDM, blaOXA-1, blaTEM-1, dfrA14, QnrB17, catII, determinant-of-bleomycin-resistance, and sul2. Novel arrangements of insertion sequences were observed in E. hormaechei plasmids. The phenotypic resistance of all the isolates matched the genotypic MDR profiles, including resistance to chloramphenicol, gentamicin, tetracycline, and cotrimoxazole.
ONT WGS with Galaxy and KNIME processing may be a feasible option for AMR surveillance in resource-limited LMICs. To the best of our knowledge, this is the first in-house whole-genome analysis workflow in the country tailored for AMR surveillance. The presence of potentially pathogenic high-MIC, MDR Enterobacter spp. with wide resistomes, including the blaNDM gene, emphasizes the urgent need to address AMR in animal production facilities within a one-health framework.
Antimicrobial resistance (AMR) is a major global health concern with a disproportionate burden on lower- and middle-income countries (LMICs). It is considered a silent pandemic and currently causes more than one million deaths annually [1]. The current death toll is predicted to rise to 10 million by 2050 if vigorous actions are not taken to curb the emergence and transmission of AMR [2]. Nearly 70% of antimicrobial drugs sold globally are used on farm animals for therapeutic, prophylactic, and growth-promoting purposes [3, 4]. Consequently, farm effluents such as feces and wastewater are major sources of antimicrobial residues and potential elevators of antibiotic-resistant bacteria (ARB) and antibiotic resistance genes (ARGs) [5]. These effluents contaminate the farm environment as well as the surrounding agricultural lands and natural waterways. Additionally, the use of manure and farm waste as fertilizer, which is a common practice in LMICs, directly contributes to the further spread of ARB and ARG [6, 7]. Furthermore, certain ARBs may persist in agricultural soils for several years after the cessation of manure use and contaminate vegetables grown on these lands [8, 9]. Therefore, the emergence and transmission of AMR in animal production systems is a prime cause driving the global AMR burden.
Many countries, including Sri Lanka, have developed national action plans (NAPs) for AMR that include AMR surveillance programs [10, 11]. However, most surveillance programs in NAPs focus on the detection of AMR in the human health sector, with concurrent lower financial inputs for surveys in the animal health sector. Even the few active surveillance programs for animals are conducted mainly for food safety purposes [11]. Active surveillance of AMR in animal production facilities is constrained by a lack of regulations and guidelines, harmonized sampling protocols, and cost-effective and robust technologies. Most currently used analytical methods are focused on the detection of ARB/ARGs in selectively cultured bacteria, such as Escherichia coli [12, 13]. Importantly, many microbial species, particularly those that naturally inhabit animal production facilities and environments that contribute to the spread of ARB, are nonculturable, difficult to grow, or difficult to identify via conventional biochemistry. Therefore, the true AMR burden in animal production facilities and the environment is often underestimated, particularly in LMICs.
Sri Lanka is a tropical South Asian LMIC. The dairy industry is the second largest animal production sector in the country, and consequently consumes a large quantity of antibiotics sold [14,15,16,17,18]. The Central Province in the up-country agroclimatic zone has the highest cattle density in the country and produces ~ 30% of the total milk output [16, 19]. The current annual milk production of Sri Lanka is approximately 480 million liters, which meets only 40% of the domestic demand. Accordingly, the import of dairy products is a major burden on the Sri Lankan economy [15, 18]. In this context, the government of Sri Lanka currently has a policy to achieve self-sufficiency in dairy products in the near future and has recently introduced many incentives to develop the dairy sector. Despite government support, AMR in the dairy sector is a major challenge that significantly increases the cost of production in addition to the inherent environmental contamination and risk to human health [20].
Despite having an NAP on AMR, Sri Lanka lacks an active AMR surveillance program for the veterinary sector and, specifically, does not have specific regulations/guidelines that mandate the surveillance of AMR in animal production facilities and the environment. Therefore, the limited information available on AMR in animal production facilities and the environment is from ad hoc surveys, which are generated by time-consuming and relatively expensive culture-dependent methods.
The development of Oxford Nanopore Technology (ONT) real-time long-read sequencing is bringing significant advantages to microbial genomics. Unlike short-read sequencing, ONT enables the identification of complex genomic features like repeat elements, structural variants, and plasmids. These capabilities enhance the accurate bacterial species identification and their corresponding AMR determinants [21, 22]. Furthermore, ONT's portability, speed, and scalability make it an attractive tool in both field and clinical settings for pathogen surveillance, outbreak response, and AMR monitoring [23, 24]. As recent studies mentioned, Thorpe et al. (2024) showed a surveillance study, how multidrug-resistant tuberculosis was readily tracked using ONT's genome and plasmid assembling capabilities compared to Illumina sequencing [25]. Schmidt et al. (2016) utilized ONT metagenomics to detect bacterial pathogens and resistance genes in clinical samples [26]. Consequently, ONT sequencing is a successful method for the detection of bacterial species and AMR surveillance.
Therefore, the objective of the present study was to evaluate a rapid, low-cost, high-throughput screening system based on next-generation sequencing coupled with open-source Galaxy software and the Konstanze Information Miner (KNIME) platform as a pilot study to screen ARBs in dairy farms in the Central Province of Sri Lanka.
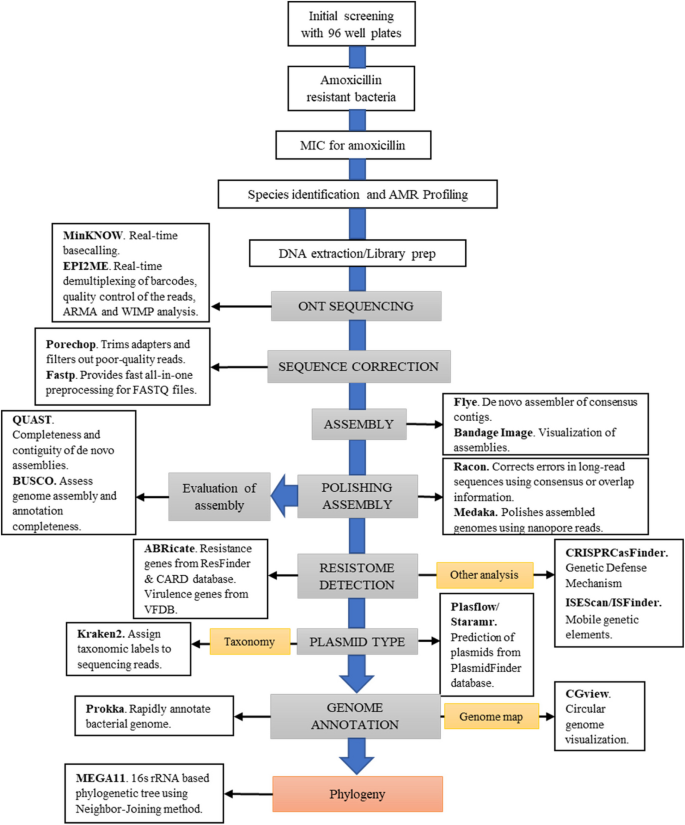
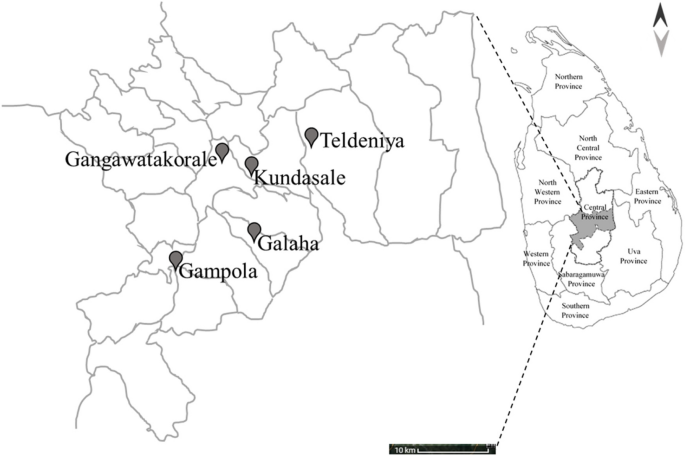
A schematic overview of the overall protocol is provided to illustrate the sequence of methodologies employed (Fig. 1). A total of 15 samples were collected from all veterinary ranges across three farms in each area, out of which four single isolates were selected for downstream genomic analysis. The geographical locations of the sampling sites are shown in Fig. 2.
The workflow for systematic analysis and visualization of multidrug-resistant (MDR) bacterial isolates. This workflow provides a systematic approach to characterize MDR bacterial isolates, incorporating rapid library preparation with transposase-based barcoding and adapter ligation via MinION sequencing. This approach facilitated the inclusion of circular genomic DNA for sequencing. Multiple rounds of assembly polishing were performed via Racon and Medaka to increase genome accuracy and minimize pseudogene artifacts. The workflow enabled a thorough characterization of genomic features, including antimicrobial resistance genes, virulence factors, and other relevant elements, providing insights into the genetic architecture of MDR bacteria. Individual bioinformatic software at each step is shown in the boxes with their main function
Geographical distribution of veterinary sampling sites in the Kandy district, Central Province, Sri Lanka. The black pins indicate the locations of the veterinary sampling sites (Gangawatakorale, Kundasale, Teldeniya, Gampola, and Galaha). The inset map highlights the location of the Central Province within Sri Lanka. The scale bar indicates a distance of 10 km for the spatial reference of the veterinary sampling sites in the Kandy district, and the arrows represent cardinal directions for orientation (north and south)
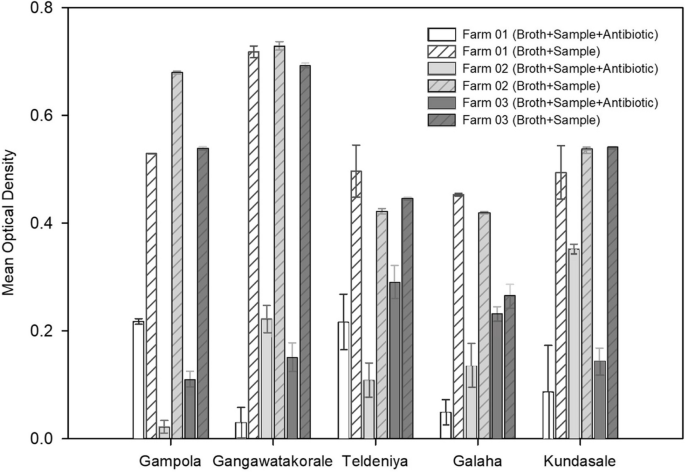
In the initial 96-well plate screening assay, all 15 cow dung samples tested showed an increase in turbidity in the wells with amoxicillin, indicating the presence of culturable, aerobic bacteria with the ability to grow in the presence of amoxicillin at a 50 µg/mL concentration in the composite samples obtained from all farms.
Four samples grown with antibiotics, at least one per veterinary range, presented an optical density (OD) > 0.2. At least one sample grown with antibiotics from the Galaha, Teldeniya, and Kundasale ranges showed OD > 50% compared with the OD of the control, indicating the presence of many amoxicillin-resistant bacteria in the samples originating from these farms (Fig. 3).
Optical density (OD) of cow dung samples from fifteen farms in Kandy. OD of the broth in the 96-well plates containing cow dung samples from 15 selected farms in the Kandy district (3 separate farms from each zone) after incubation for 24 h in the presence of amoxicillin (50 μg/mL) and without amoxicillin. The results are presented as the mean ± SEs (n = 4). A zero value was obtained for the OD in the negative control with only broth. Graphs were constructed via SigmaPlot V.11.0 [27]
Growth was observed (> 0.2 OD value in the initial 96-well plate assay compared to the control) in all samples plated on single culture plates with amoxicillin (50 μg/mL), confirming the presence of culturable, aerobic, amoxicillin-resistant bacteria on all farms. At least three isolates per farm were randomly selected for the MIC assays.
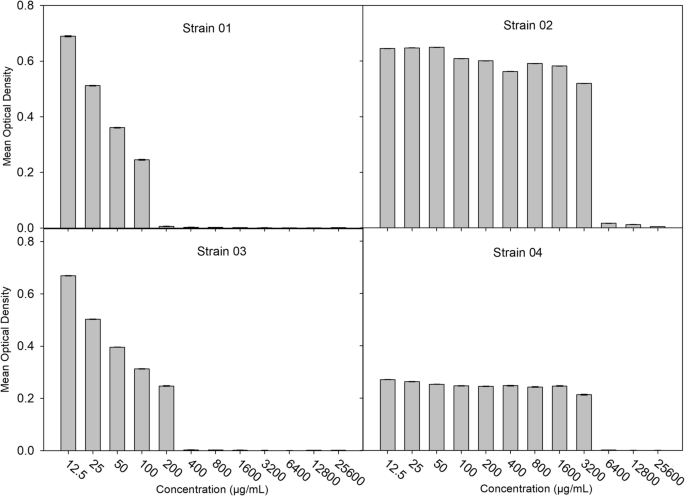
The observed MIC values for amoxicillin ranged from 12.5 µg/mL to 3200 µg/mL. Four isolates presented MICs ≥ 100 µg/mL (Fig. 4). The additional data file shows the mean OD values for samples from each veterinary range and all the MIC values obtained for selected isolates (see Additional file 1).
Four bacterial isolates displayed amoxicillin resistance with MICs ≥ 100 µg/mL at an OD of 620 nm. The results are presented as the mean ± SEs (n = 4), and the MIC breakpoint for amoxicillin is 8 µg/mL according to EUCST 2022. Isolates IDs are Strain 01, Strain 02, Strain 03, and Strain 04. Graphs were constructed via SigmaPlot V.11.0 [27]
Based on the initial 96-well plate assay (OD > 0.2) and minimum inhibitory concentration threshold (MIC ≥ 100 µg/mL amoxicillin), four isolates were selected for this AMR study, which originated from four different farms. (Strain 01 from Gangawatakorale farm 02, Strain 02 from Theldeniya farm 03, Strain 03 from Kundasale farm 02, Strain 04 from Galaha farm 03).
All four isolates that presented the highest MIC for amoxicillin (≥ 100 µg/ml) were gram-negative motile bacilli. All were negative for indole, methyl red, and Voges–Proskauer tests and did not produce hydrogen sulfide. All the isolates were positive for citrate and urease and fermented lactose, sucrose, mannitol, and xylose, suggesting that the isolates may be identified as Enterobacter cloacae complex. In addition to amoxicillin resistance, these four isolates were resistant to ciprofloxacin and chloramphenicol. Additionally, one isolate was resistant to tetracycline. In contrast, the remaining 03 isolates were resistant to gentamycin and cotrimoxazole. Accordingly, all the isolates were phenotypically categorized as MDR (Table 1).
The extracted DNA from the above four isolates was confirmed to be of high molecular weight with fragment sizes exceeding 10 kb by agarose gel electrophoresis (Supplementary Figs. 1 & 2). Purity assessment via a Nanodrop spectrophotometer revealed OD 260 nm/280 nm ratios ranging from 1.86 to 1.94, indicating minimal protein contamination (Additional file 2).
MinION DNA sequencing generated > Q7 525.2 Mb (107 ×) for Strain 01, 398.1 Mb (81 ×) for Strain 02, 180.2 Mb (36 ×) for Strain 03, and 339.2 Mb (69 ×) for Strain 04. These coverage values mentioned raw read coverage, while assembly coverage generated by Flye is detailed in Table 2. Across the assembly tools, the chromosome sequences commonly appeared as circularized 4.8–4.9 Mb contigs, and plasmids ranged between 8 and 171 kb except for Strain 01, which lacked plasmids (Table 2). Each of the four isolates had circular genome configurations. At least one mega-plasmid, defined as a plasmid > 170 kb, was detected in three isolates (Table 2), with the exception of Strain 01. These isolates commonly harbor the replicons IncFIB (pECLA) and IncFII (pECLA). The IncX3 replicon microplasmid (< 53.7 kb) and unnamed (without a replicon) microplasmid (> 9 kb) were present separately in Strains 02, 03, and 04. After sequence correction and assembly polishing, Strains 01, 02, 03, and 04 presented average coverage values of 106x, 75x, 35x, and 66x, respectively. A summary of assembly quality is provided (Additional file 3).
According to the annotation results (Table 3, Additional file 11), Strain 01 presented 5052 coding DNA sequences (CDS: including hypothetical proteins and putative genes) with a total of 5164 genes, alongside 25 ribosomal RNAs (rRNAs), 86 transfer RNAs (tRNAs), and 1 transfer-messenger RNA (tmRNA). Strain 02 presented 5544 genes, along with 5433 CDSs, 25 rRNAs, 85 tRNAs, and 1 tmRNA. Strain 03 included 6014 genes, with 5904 CDSs, 25 rRNAs, 84 tRNAs, and 1 tmRNA. Strain 04 included 5669 genes alongside 5558 CDSs, 25 rRNAs, 85 tRNAs, and 1 tmRNA. These results, detailed in Supplementary file 11, highlight variations in gene content with an E value cutoff of 1e-06 among the isolates while maintaining the consistent presence of ribosomal and transfer RNA elements across all isolates.
As virulence genes, ompA in the chromosomes of all studied isolates and csgG in Strain 01 and entB in Strains 02, 03, and 04 were identified with coverage values greater than 99% (Additional file 8). The Enterobacter isolates had low-confidence (evidence level 1) CRISPR arrays without associated Cas genes. All isolates had one to three arrays, located on chromosomes and/or mega-plasmids, with variable repeat lengths (23–48 bp). The arrays had unique spacer sequences (Supplementary Figs. 8, 9, 10, & 11 and Additional file 9).
Analysis of insertion sequences (ISs) annotated by ISEScan and ISFinder in four isolates studied revealed the presence of 11 different IS families, including a novel IS (Table 4, Additional file 10). Each isolate’s chromosome exhibited 2 distinct IS families: IS4 and IS481 in Strain 01 and IS1 and IS3 in the other three isolates. Furthermore, the mega-plasmids of Strains 02, 03, and 04 consistently contained seven IS families each, including the novel IS. Similarly, the microplasmids across these isolates consistently harbored four IS families, which also included the novel IS. All identified IS clusters and related details, including terminal inverted repeats, are provided in additional file 10, and locations are clearly shown in Supplementary Figs. 8, 9, 10, & 11. This comprehensive analysis highlights the presence and distribution of IS elements, emphasizing both consistency and variability across different genetic elements in the studied isolates. Furthermore, two transposon (Tn) elements with more than 90% query coverage were found in Strains 02, 03, and 04 according to the ISFinder BLAST results.
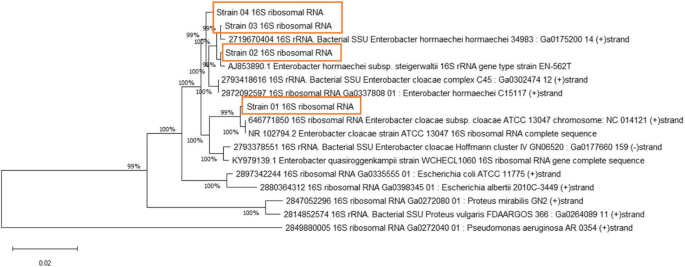
Taxonomic assignment via the WIMP (What’s In My Pot) feature of the EPI2ME identified the following taxa: 100% Enterobacteriaceae, which represents 100% E. cloacae for Strain 01; and Enterobacter hormaechei, which represents Strains 02 (92%), 3 (77%) and 4 (72%) with threshold value ≥ 750000 (Additional file 4, Supplementary Fig. 3). In a subsequent test from Kraken2, classification results were obtained for Strain 01, namely, Enterobacter cloacae subsp. cloacae ATCC 13047 (taxid 716541), and for the other three isolates, namely, Enterobacter hormaechei subsp. xiangfangensis (taxid 1296536) (Additional file 5), from their chromosomes. Additionally, NCBI BLAST searches of the complete sequences of the mega-plasmids in Strains 02, 03, and 04 revealed that E. hormaechei presented > 99% identity (Additional file 6). To visualize these findings, a 16S rRNA-based phylogenetic tree was constructed (Fig. 5). Additional data files show the taxonomic assignment and genome maps (see Additional files 4, 5, & 6). The GTDB-Tk results provided improved taxonomic resolution and confirmed the species-level assignments of our isolates. Detailed summary of the classification output has included in the Additional file 5.
Phylogenetic tree based on 16S rRNA sequences of four sequenced Enterobacter isolates. The phylogenetic tree was inferred via the neighbor‒joining method [28], and the optimal tree is shown. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) is shown next to the branches [29]. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances (number of nucleotide substitutions per site) were computed via the Kimura-2-parameter method [30]. All ambiguous positions were removed for each sequence pair. There was a total of 1519 positions in the final dataset. Evolutionary analyses were conducted in MEGA11 [31]
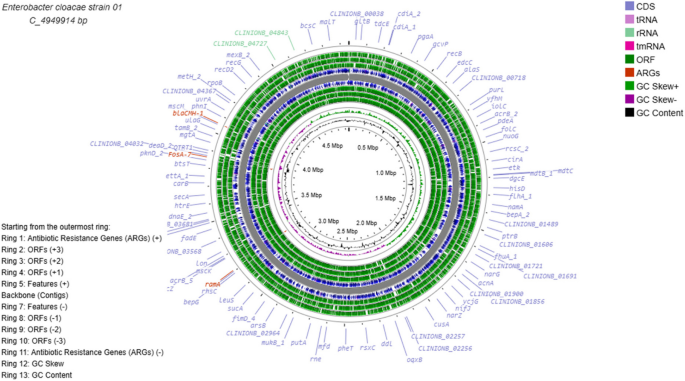
The chromosome sizes ranged from 4.8–4.9 Mb. Notably, the resistance genes ramA, blaCMH-1, and fosA_7 were present in Strain 01 (Fig. 6), conferring resistance to β-lactams, fluoroquinolones, phenicols, rifamycins, tetracyclines, triclosan, and fosfomycin antibiotic classes. In Strains 02, 03, and 04, the β-lactam resistance gene (blaACT) was present in the chromosome. Notably, all the other resistance genes (≥ 92%) were present on plasmids within these three isolates (Figs. 6 & 7 and Supplementary Figs. 4, 5, 6, & 7). Further details of the identified resistance genes from ABRicate are shown in Additional file 7.
Genome map of Strain 01. The bacterial chromosome is 4.9 Mb in size. Antibiotic resistance genes are highlighted in red. The circular map was constructed by uploading the Galaxy annotated GenBank file into the CGView/Proksee genome visualization tool
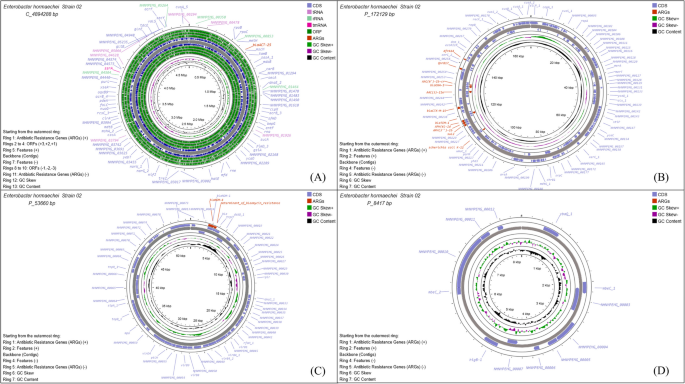
Genome map of Strain 02. Panel A shows the 4.8 Mb bacterial chromosome, while Panels B – D display plasmids ranging in size from 8 to 171 kb. Antibiotic resistance genes are highlighted in red. The circular maps were generated by uploading the annotated GenBank file (from Galaxy) into the CGView/Proksee genome visualization tool
The IncX3 replicon microplasmid contained resistance genes of blaNDM-4 and determinant_of_bleomycin_resistance, which confer resistance toward β-lactams and glycopeptides, respectively. A double-replicon-identified mega-plasmid contained aminoglycoside, β-lactam, diaminopyrimidine, fluoroquinolone, phenicol and sulfonamide resistance (AAC(3)-IIe, AAC(6’)-Ib-cr, APH(3'')-Ib, APH(6)-Id, blaCTX-M-15, blaOXA-1, blaTEM-1, dfrA14, QnrB17, catII & sul2) (Table 2, Additional file 7). The extended-spectrum β-lactamases BlaTEM and blaCTX-M have been detected in Strains 02, 03, and 04 as intrinsic resistance genes. Additionally, the blaACT, blaNDM, blaOXA (Strains 02, 03, and 04), and blaCMH (Strain 01) genes have also been identified as intrinsic beta-lactam resistance genes.
In the comparative analysis of AMR gene predictions from the Comprehensive Antibiotic Resistance Database (CARD) and ResFinder databases from ABRicate across studied isolates (Additional file 7), all genes except one in each isolate were predicted, as marked (c) in Table 2. Specifically, the ramA gene was not detected in Strain 01 via ResFinder, whereas it was predicted via the CARD database. Additionally, the fosA gene was identified in Strain 01 via ResFinder with > 96% identity. Furthermore, the novel determinant_for_bleomycin_resistance gene was predicted in Strains 02, 03, and 04 via the CARD database but was not identified via ResFinder.
The multidrug-resistant Enterobacter cloacae complex (ECC), which represents ESKAPE pathogens (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter spp.), which shows increased resistance to commonly used antibiotics, is emerging as a global threat [32]. In addition to intrinsic β-lactam resistance genes, members of the ECC family acquire multiple classes of antibiotic resistance genes. The misuse and overuse of antibiotics in livestock farms is one of the major contributors to the development of AMR, including multidrug-resistant ECC. The significance of animal farming in regions with limited resources cannot be underestimated, particularly as many countries shift toward more intensive animal husbandry practices. This shift, unless coupled with good husbandry practices and increased hygiene, results in increased usage of antimicrobial agents, consequently increasing the potential for increased exposure to AMR for both animals and humans on a global scale [33].
Compared with traditional agar plate-based screening, the 96-well plate screening method developed in this study can be easily used to screen multiple samples at once. Furthermore, the identification of specific resistant bacteria or groups of bacteria via the use of different antibiotics for selection can potentially be adaptable [34]. The ONT MinION portable sequencing platform coupled with an open-source workflow provides a feasible solution for genomic analysis, especially in resource-constrained environments, remote locations, and onsite applications [35]. Compared with conventional short-read technologies, the long-read capacity of ONT provides de novo assembly by spanning complex regions such as structural variants and repetitive regions to create accurate assemblies [36, 37]. This technology is particularly advantageous for resolving plasmids as single contigs [38], enabling the accurate identification of clinically relevant genes, including AMR determinants, virulence factors, and MGEs, thereby providing robust and reliable insights for AMR surveillance and other predictions [37, 39]. Additionally, we elected to use the ONT rapid chemistry platform because of the unique transposase-mediated cutting of chromosome and plasmid DNA and attachment of adapters, resulting in 100% MDR detection in chromosomes and plasmids. Notably, to the best of our knowledge, this marks the first instance of an in-house-developed full pipeline covering bench work to bioinformatics in Sri Lanka for AMR surveillance.
The intrinsic resistance of Enterobacter species is coordinated by key genetic elements, shedding light on their importance and the alarming challenges they pose. In this context, the acrAB gene, encoding an efflux pump that functions as a molecular bouncer, actively removes a diverse array of antibiotics from bacterial cells, and this process is regulated by local regulators such as acrR and global regulators such as marA, soxS, and ramA [40, 41]. Notably, our study revealed the presence of the ramA gene within the chromosome of Strain 01. CARD-based ABRicate analysis detected the chromosomal ramA gene in Strain 01, which encodes a regulatory gene involved in multidrug resistance mechanisms. These intrinsic traits may not always be flagged by the ResFinder database alone, which is primarily curated to detect acquired resistance genes, typically plasmid-borne or horizontally transferred genes.
Furthermore, the presence of beta-lactam resistance determinant genes, including blaCMH-1, blaCTX-M-15, blaACT-25, blaNDM-4, blaOXA-1, and blaTEM-1, in these isolates aligns with previous findings [42, 43]. Extended-spectrum beta-lactamases (ESBLs) are a group of beta-lactamase enzymes that confer resistance to beta-lactams, including penicillin; first-, second-, and third-generation cephalosporins; and monobactams (e.g., aztreonam) [44,45,46]. The presence of ESBLs is associated with the expression of SHV, TEM, and CTX-M enzymes encoded by some blaSHV variants (e.g., SHV-2, SHV-5), blaTEM variants (e.g., TEM-4, TEM-5, TEM-10, TEM-12, TEM-30), and blaCTX-M genes, respectively [44, 46]. The blaCTX-M-15 gene, which is the most widespread and clinically significant subgroup of ESBLs [47, 48], was identified in Strains 2, 3, and 4 in this study. ESBL genes can be commonly found in both the chromosomes and plasmids of species within the Enterobacteriaceae family [44], and in our study, the blaCTX-M-15 gene is located on plasmids, which can be easily transferred between bacteria, leading to the rapid spread of resistance. The rise of ESBL-producing genes is associated with widespread resistance to multiple drugs, presenting a significant obstacle in the treatment of infections [48]. In clinical settings, particularly in the human context, the finding of carbapenem resistance is the most concerning aspect [49]. Identifying such specific genes via in-depth genomic analysis, such as nanopore sequencing, has a significant advantage over routine PCR [26].
The World Health Organization (WHO) has published a list of the most threatening bacteria with emergent AMR, categorizing them into 3 priority groups on the basis of urgency for new antibiotics: critical, high, and medium [50]. Among these, Carbapenem-resistant Enterobacterales (CRE) are classified within critical groups of priority [51]. CRE has been reported in various countries, including Spain, Australia, the United States, India, Korea, and China [52]. Among the three types of carbapenemases, the emergence of Ambler class B New Delhi metallo-β-lactamase (blaNDM-1) is a critical issue worldwide [53]. The detection of IncX3 plasmid-mediated blaNDM-4 and blaNDM-15, which are variants of blaNDM-1, in this study highlights the spread of the blaNDM gene within Sri Lankan dairy farms. This finding suggests the potential of blaNDM-carrying plasmids or bacteria to be transmitted from livestock to the environment via manure [54, 55].
Enterobacter species exhibit resistance to quinolones through mutations in gyrA and parC, which encode subunits of DNA gyrase and topoisomerase IV [56]. These alterations disrupt the binding of quinolone antibiotics, subsequently reducing their efficacy [57]. Plasmid-mediated resistance was observed in our study, particularly with the identification of QnrB17, which contributes to fluoroquinolone resistance. Because Qnr genes alone are known to confer only low-level resistance, annotation using Prokka identified both gyrA and parC genes within our assemblies without mutations of gyrA [codons 83 (Ser83) and 87 (Asp87)] and parC [codons 80 (Ser80) or 84 (Glu84)] genes. This suggests that quinolone resistance is genetically unlikely in these isolates.
Additionally, aminoglycoside resistance mechanisms, including genes such as AAC(3)-IIe and AAC(6')-Ib-cr, which encode enzymes capable of modifying aminoglycosides and rendering them ineffective against bacterial resistance, have been identified [57]. Resistance to aminoglycosides in Enterobacteriaceae commonly results from mobile genes encoding aminoglycoside-modifying enzymes (AMEs) that modify sugar moieties, leading to aminoglycoside inactivation [58, 59]. Both aac (acetylation) and aph (phosphorylation) involve this type of modification [58]. Our study further identified genes encoding AMEs, APH(3'')-I, APH(6)-Id, AAC(3)-IIe, and AAC(6')-Ib-cr.
Apart from the findings of intrinsic resistance genes in Enterobacteriaceae, the presence of genes such as catII (phenicol resistance gene), dfrA14 (diaminopyrimidine), determinant of bleomycin resistance, and sul2 (sulfonamide) in Strain 02—04 could be attributed to the possibility that these Enterobacter spp. acquired resistance genes from other microorganisms. This is further confirmed by the discovery of foreign genetic elements, including ISs, Tn, and plasmids, which are indicative of the potential for horizontal gene transfer and the acquisition of resistance genes in these isolates [43, 60, 61].
Comparative findings of ARGs highlight the differences in performance between ResFinder and CARD, particularly in detecting resistance genes with the novelty and specificity of the studied isolates. ResFinder is generally considered more accurate when detecting well-characterized, clinically relevant, or mobile ARGs [62]. The CARD was chosen since it is a more comprehensive database than ResFinder and contains some genes that are not clinically relevant and/or mobile and are continuously updated with newly discovered resistance mechanisms [63, 64].
The common structure of the CRISPR/Cas system consists of a CRISPR array, Cas genes, and leader regions. These isolates have only CRISPR arrays, which suggests that they may have a nonfunctional or putative CRISPR/Cas system [65]. According to Medina-Aparicio et al. [66], the distribution of the CRISPR/Cas system in the Enterobacteriaceae family is not regular and rarely occurs in the genus Enterobacter. The variation in CRISPR sequences among the different species highlights genetic diversity within the Enterobacter species, which could reflect differing evolutionary histories and environmental pressures.
Mobile genetic elements (MGEs) are frequently situated on these isolates, and almost all ARGs are situated within the ISs (Additional file 10 and Supplementary Figs. 8, 9, 10, & 11). Furthermore, most of the ISs identified (IS110, IS1380, IS3, IS5, IS6, IS30, and ISNCY), with the exception of the novel ISs, have previously been documented in plasmids from enteric bacterial species using In silico analysis and the ESKAPE group of organisms [43, 67]. Notably, a key player in the evolution of MDR plasmids and chromosomal islands in Enterobacteriaceae is the Tn3-type transposon, as highlighted in prior studies [68].
Plasmids are categorized into incompatibility (Inc) groups on the basis of their ability to coexist within the same bacterial isolate. Incompatibility arises when two plasmids with the same replication machinery and control systems are present in the same cell, interfering with their replication and maintenance [69]. Plasmids within the same Inc. group are therefore incompatible and cannot coexist in a single host. Among Enterobacterales, twenty-seven major plasmid incompatibility groups are associated with ARGs, with IncF, IncA/C, and IncX plasmids being the most prevalent in carbapenemase production [70]. Incompatibility arises only when separate plasmids with the same replicon types attempt to coexist within the same host cell. In our study, we identified plasmids without multiplicity within each isolate according to the coverage of the assembly (Additional file 3). Therefore, the identification of a multireplicon IncF plasmid alongside a separate IncX3 plasmid within the same bacterial isolate in our study suggests that these plasmids rely on different mechanisms of replication and maintenance. This allows them to coexist without triggering incompatibility, as their replication systems do not interfere with one another. The coexistence of these plasmids within the same cell highlights their compatibility, enabling the stable maintenance of both plasmids and the potential for efficient dissemination of antimicrobial resistance genes. Moreover, we identified plasmids containing two replicons, IncFIB (pECLA) and IncFII (pECLA), within the same plasmid. The coexistence of these replicons on a single plasmid does not trigger incompatibility because they are integrated into the same DNA molecule, forming a multireplicon plasmid. Notably, certain transmissible plasmids, particularly those classified into IncI and IncF groups, have been previously shown to play pivotal roles in facilitating the transmission of resistance [68]. We identified IncF plasmids in our study. Additionally, IncX3 plasmids, which are primarily found in Enterobacteriaceae, are characterized by their narrow host range. They exhibit notable features such as high conjugation ability, high stability, and negligible fitness costs [71]. According to Wang et al., IncX3 plasmids can be efficiently transferred to multiple Enterobacterial species at frequencies similar to or higher than those of their studied IncFII plasmid-carrying blaCTX-M isolate [72].
In addition to these findings, the whole genomes of all the isolates contained 25–31% hypothetical proteins out of the total CDS. As described in the literature, nearly 30% to 40% of genes of most bacterial genomes are classified as unknown or hypothetical in several studies [73,74,75]. Additionally, genome-level comparison of strains 2–4 using D-GENIES dot plot analysis to assess the extent of sequence similarity revealed a high degree of genomic similarity, with > 75% of the sequences aligning at an identity of more than 99%, indicating that these genomes are nearly identical.
Although portable sequencing technologies such as ONT and open-source workflow platforms are becoming more widely available, resource-constrained settings and LMICs in general still face many obstacles. The main limitations in our study were the difficulty in obtaining third-party consumables from a single supplier, the lengthy documentation required for customs clearance, the maintenance of the cold chain for ONT supplies from the entry airport to the end-user facility, and slow internet speed. Some ONT supplies require refrigeration, whereas the rest are shipped frozen. This adds a substantial premium on shipping and handling costs. Notably, accurate quantification of very high-quality genomic DNA is critical to ensure sufficient depth of coverage to achieve the desired results. This is a major challenge in our setting, as commercial DNA extraction kits are very expensive, and the extraction and quantification process require substantial laboratory support. Similarly, an uninterrupted power supply is critical for the entire WGS run. While this study did not systematically capture detailed antibiotic usage practices at different farms, integrating such details in future investigations would strengthen the reliability and stability of the observed relationship between antimicrobial use and the abundance of AMR species at each farm, particularly high-risk groups such as ESKAPE pathogens. Also, the small sample size used in this study may have limited the potential for detecting a wider range of species. A bigger sample size, particularly from additional farms, could have provided higher statistical power, revealing additional species and making the study more robust. Moreover, long-read-only sample assembly might allow artifacts to build up the genome in the wrong way with annotation results. This approach would support more targeted and evidence-based interventions. Overall, we were able to develop a rapid screening platform comprising in-house WGS coupled with comprehensive genomic analyses employing scalable and cost-effective solutions that can be used for AMR surveillance in resource-limited settings such as LMICs.
Oxford Nanopore WGS with open-source Galaxy software and KNIME analytics platform processing is a potentially feasible option for AMR surveillance in resource-limited LMICs. This approach generated high-quality genomic data, rapidly identified the presence of the ESKAPE pathogen Enterobacter spp., and captured the entire resistome using a minimum sample number. To the best of our knowledge, this is the first in-house whole-genome analysis workflow in the country tailored for AMR surveillance. The presence of potentially pathogenic high-MIC, MDR Enterobacter spp. with wide resistomes, including the blaNDM gene, emphasizes the urgent need to address AMR in animal production facilities within a one-health framework. The identification of carbapenem resistance genes such as blaNDM, virulence genes, and MGEs, including novel arrangements of insertion sequences, shows the ability of WGS to capture evolutionary and epidemiological trends in AMR spread.
Fifteen farms from five veterinary ranges (three farms per range) in the Kandy district of the Central Province (Kundasale, Teldeniya, Galaha, Gangawatakorale, and Gampola) were selected on the basis of the recommendation of the Provincial Director of Veterinary Services. Three small-scale dairy farms within each range were selected for convenient sampling using sterile disposable gloves in clean plastic bags. Composite dung samples were collected from each farm. Approximately 300 g of fresh dung samples were collected from six locations from each farm, combined, and thoroughly mixed, and approximately 100 g of the composite sample was collected into sterile polythene bags and transported to the laboratory on ice.
Initial screening was conducted on a 96-well plate-based assay in the presence of amoxicillin. The amoxicillin/penicillin group is the most commonly used antibiotic on dairy farms in Sri Lanka [20, 76].
Composite dung samples were thoroughly mixed, 1 g was suspended in 10 mL of sterile water, and tenfold serial dilutions were prepared using sterile water. In the pilot study, 25 µL of 10–2 diluted dung suspension was added to quadruplicate wells of 96-well plates containing 100 µL of nutrient broth with and without 50 µg/mL amoxicillin (Sterling Lab, India). Quadruplicate wells without samples and without antibiotics were used as sterility controls.
After the plate was incubated for 24 h at room temperature, the optical density was measured via a microplate reader (Thermo Scientific Multiskan FC) at 620 nm.
The samples that had grown in the presence of amoxicillin in the 96-well plate assays were inoculated onto nutrient agar plates containing 50 µg/mL amoxicillin and incubated at 37 °C for 24 h. The isolated colonies were subcultured onto nutrient agar plates to obtain pure cultures. The inoculum was prepared from the pure cultures by suspending the isolated colonies in sterile distilled water, and the concentration was adjusted to 0.5 McFarland standard. The MIC for amoxicillin was determined via the micro broth dilution method [77, 78].
The isolates with the highest MICs for amoxicillin were partially characterized via Gram staining, motility testing, and conventional biochemical methods (indole, methyl red, Voges–Proskauer, citrate, urease, hydrogen sulfide, lactose, sucrose, mannitol and xylose fermentation according to standard protocols) [79]. The isolates with the highest MICs for amoxicillin were further characterized by determining the AMR for multiple drug classes of amoxicillin [10 µg (β lactams)], ciprofloxacin [5 µg (quinolone)], chloramphenicol [30 µg (phenicols)], gentamicin [10 µg (aminoglycoside)], tetracycline (30 µg) and trimethoprim/sulfamethoxazole (25 µg)] via the Kirby–Bauer disk diffusion method according to EUCAST 2022 guidelines.
DNA was extracted from 10 mL of overnight (37 °C) nutrient agar broth cultures using the QIAamp DNA Mini Kit (Qiagen: Chadstone, Victoria, Australia) according to the manufacturer’s instructions. The purity and concentration of the DNA were measured via a Nanodrop ND-2000—OD 260 nm/280 nm of 1.8 and OD 260 nm/230 nm of 2.0–2.2. The average fragment size greater than 10 kb was analyzed via a low percentage agarose gel (see Additional file 12).
The MinION sequencing libraries with 400 ng of DNA input each were prepared with the Rapid Barcoding Sequencing Kit (SQK-RBK004). Briefly, 7.5 μl of 400 ng of template DNA was mixed with 2.5 μl of Fragmentation Mix RB01-12, and the tube was incubated at 30 °C for 1 min and then at 80 °C for 1 min. All the barcoded samples were pooled, and 1 µl of Rapid Adapter (RAP) was added to 10 µl of barcoded DNA, followed by incubation for 5 min at room temperature. The sequencing was performed in a portable MinION sequencing device with MinION R9.4.1 flow cells. Sequencing was started with MinKNOW™ software, and was used to perform base-calling and barcode demultiplexing during sequencing and through post run analysis. The generated FASTQ-pass reads were sent through the EPI2ME workflow to align the sequences and identify antibiotic resistance genes in real time. Reagents, flow cells, MinION sequencing device MinKNOW™ and EPI2ME software were sourced through ONT (Littlemore, Oxford, United Kingdom), and all sequencing and data analyses were carried out in-house.
Primary acquisition of data and real-time base calling were carried out via the graphical user interface MinKNOW™, v. 4.5.5. The demultiplexing of barcodes, quality control of the reads, and assignment of taxonomy via WIMP were also accomplished in real time via the EPI2ME platform. All quality reads (quality score above 8) were extracted after 23 h and 10 min of sequencing for downstream analysis. The WIMP results were subsequently analyzed by assigning a threshold value of ≥ 750,000 scores via KNIME analytics platform (Supplementary Fig. 12).
Galaxy bioinformatics tools were used to assemble the genome with ONT reads (https://usegalaxy.org/) (the complete workflow in Galaxy is shown in Supplementary Fig. 13). Preprocessing of FASTQ files was conducted via Porechop v. 0.2.4 [80] and fastp v. 0.23.4 [81, 82] with default parameters. De novo assembly was performed via Flye v. 2.9 [83, 84] by specifying the input as Nanopore reads-Nanopore corrected to a minimum coverage of ~ 30X, and Bandage v.0.8.1 [85] was used to visualize the assemblies. Further polishing and error correction of the genome assemblies were conducted via Racon v. 1.5.0 with default parameters and Medaka v. 1.12.1 [86, 87] by specifying the final polishing with the model r941_min_fast_g507. The completeness and contiguity of the de novo assemblies were evaluated via QUAST v. 5.0.2 [88, 89] and BUSCO v. 5.5.0 [90]. The locations of the acquired antibiotic resistance genes were determined via ABRicate v. 1.0.1 from ResFinder and CARD [91]. The genomes were annotated via the rapid annotation software Prokka v. 1.14.6, which provided a list of genes [92, 93]. ONT FASTQ sequencing data have been deposited at the National Center for Biotechnology Information (NCBI) under Bioproject ID PRJNA1102716 in the Sequence Read Archive (SRA) (www.ncbi.nlm.nih.gov/sra/). The corresponding accession numbers are as follows: Strain 01 (SRR28762089), Strain 02 (SRR28762092), Strain 03 (SRR28762091), and Strain 04 (SRR28762090). Additionally, assembly files and associated annotated data are also available under the above bioproject ID. The accession numbers are as follows: Strain 01 (CP165725), Strain 02 (CP171250-CP171253), Strain 03 (CP178584-CP178587), and Strain 04 (CP171246-CP171249).
Virulence genes were determined via ABRicate v. 1.0.1 VFDB (Virulence Factor Database) [91], and plasmids were identified via plasflow v.1.1.0 PlasmidFinder v. 2.1.6 and staramr v. 0.7.2 [94,95,96]. For precise k-mer alignments and taxonomic classification on the basis of genomes and plasmid databases, Kraken2 v. 2.1.3 [97] was employed. An additional taxonomic classification was performed using GTDB-Tk v2.4 with default parameters [98]. The clustered regularly interspaced short palindromic repeats (CRISPR)/Cas system was identified via CRISPRCasFinder v. 1.1.2 [99]. Subsequently, the spacer sequences were aligned against the E. cloacae and E. hormaechei genome sequences deposited in the National Center for Biotechnology Information (NCBI) GenBank via the NCBI BLAST algorithm. Resistance genes and megaplasmid contigs were subjected to BLASTn analysis against the NCBI database to discern whether resistance genes, plasmid sequences, and taxonomy identification have previously been reported. For visualization of whole-genome sequences, CGView/Proksee was employed [100]. This comprehensive approach allowed for the identification of virulence factors, plasmids, and resistance genes, along with taxonomic classification and visualization of the genomic context. MGEs were detected via ISEScan v1.7.2.3 [101], and the ISFinder [102] database (https://isfinder.biotoul.fr/) was used to identify transposable elements and their inverted repeat sequences. Additionally, we performed a genome-level comparison of strains 2–4 using D-GENIES v2 dot plot analysis to assess the extent of sequence similarity [103].
The Galaxy workflow is available at the Galaxy|Europe site (https://usegalaxy.eu/u/l.r.l.s.k/w/enterobacter-wgs-ont). All raw sequencing data, the genome assembly of Enterobacter spp., and annotated data are available at the National Center for Biotechnology Information (NCBI) under the Bioproject ID PRJNA1102716. The ONT DNA sequencing data accession numbers deposited in the SRA (www.ncbi.nlm.nih.gov/sra/) are as follows: Strain 01 (SRR28762089), Strain 02 (SRR28762092), Strain 03 (SRR28762091), and Strain 04 (SRR28762090). The genome assembly accession numbers are as follows: Strain 01 (CP165725), Strain 02 (CP171250-CP171253), Strain 03 (CP178584-CP178587), and Strain 04 (CP171246-CP171249). Additional data are provided within the supplementary information files.
- AMR:
-
Antimicrobial resistance
- ARB:
-
Antibiotic-resistant bacteria
- ONT:
-
Oxford Nanopore Technology
- MIC:
-
Minimum inhibitory concentration
- MDR:
-
Multidrug resistant
- LMICs:
-
Lower- and middle-income countries
- ARG:
-
Antibiotic resistance gene
- NAP:
-
National action plan
- EUCAST:
-
European Committee on Antimicrobial Susceptibility Testing
- DNA:
-
Deoxyribonucleic acid
- OD:
-
Optical density
- nm:
-
Nanometer
- kb:
-
Kilo base
- WIMP:
-
What’s In My Pot
- KNIME:
-
Konstanze Information Miner
- CARD:
-
Comprehensive Antibiotic Resistance Database
- Mb:
-
Megabase
- IS:
-
Insertion sequence
- NCBI:
-
National Center for Biotechnology Information
- rRNA:
-
Ribosomal RNA
- CRISPR:
-
Clustered Regularly Interspaced Short Palindromic Repeats
- Tn:
-
Transposon
- MGE:
-
Mobile genetic element
- CDS:
-
Coding DNA Sequences
- tRNA:
-
Transfer RNA
- tmRNA:
-
Transfer-Messenger RNA
- ECC:
-
Enterobacter cloacae Complex
- WGS:
-
Whole genome sequencing
- ESBLs:
-
Extended-spectrum beta-lactamases
- VFDB:
-
Virulence Factor Database
- PCR:
-
Polymerase Chain Reaction
- AMEs:
-
Aminoglycoside-modifying enzymes
- SRA:
-
Sequence Read Archive
- CRE:
-
Carbapenem-resistant Enterobacterales
The authors would like to thank the following individuals from the Faculty of Veterinary Medicine & Animal Science, University of Peradeniya. Prof. Ruchika Fernando for generously providing access to the plate reader at the Department of Veterinary Public Health & Pharmacology. Dr. Samanthika Jagoda and Dr. Pavithra Dhanapala at the Department of Veterinary Pathobiology assisted with DNA extraction. Mr. Sampath Bandara and Dr. Yashodha Basnayake from the same department provided technical support for bacterial culture and antibiotic sensitivity testing.
This study was financially supported by the Postgraduate Institute of Science, University of Peradeniya, under grant number PGIS/2020/17. The funding body did not play a role in the design, analysis, or reporting of the study.
Ethics approval was not required for sample collection, as all the samples were of environmental origin. Informed consent for sample collection was obtained from the regional director of veterinary services and the respective farm managers.
Not applicable.
The authors declare no competing interests.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
Kumari, L.S., Siriwardhana, D.M., Liyanapathirana, V. et al. Rapid whole genome sequencing for AMR surveillance in low- and middle-income countries: Oxford Nanopore Technology reveals multidrug-resistant Enterobacter cloacae complex from dairy farms in Sri Lanka. BMC Vet Res 21, 351 (2025). https://doi.org/10.1186/s12917-025-04800-1