Virology Journal volume 22, Article number: 151 (2025) Cite this article
Monkeypox virus (MPXV) infection has garnered significant global attention due to its rising incidence and substantial public health implications. A rapid, sensitive, and accurate diagnostic method is urgently required to enable early intervention and effective management of MPXV outbreaks.
In this study, we developed a novel one-step assay that integrates loop-mediated isothermal amplification (LAMP) with CRISPR/Cas12b in one-pot for the detection of MPXV. The entire detection process did not require opening the lid of the reaction tube and could be completed within 40 min using extracted viral nucleic acids, which is faster than real-time quantitative PCR (qPCR). And the results could be interpreted through either real-time fluorescence or naked-eye visualization. The limit of detection (LOD) of the assay was demonstrated to be 6.5 copies per reaction and no cross-reactivity with other pathogens such as HSV, EBV, CVA16, EV-A71, and MV was found. Furthermore, when evaluated using 113 clinical samples, the assay achieved 100% sensitivity (71/71) and 100% specificity (42/42) compared to the qPCR.
In resource-limited settings, our method requires only a portable heat block or water bath and a blue light or ultraviolet flashlight for visual detection of MPXV, making it highly accessible. The integration of LAMP and CRISPR/Cas12b provides a robust, user-friendly platform for point-of-care testing, with promising potential for the rapid molecular diagnosis of infectious diseases.
Monkeypox is a viral disease caused by the monkeypox virus (MPXV), an enveloped double-stranded DNA virus belonging to the Orthopoxvirus genus within the Poxviridae family. The disease is primarily transmitted through close contact with infected animals or individuals and is characterized by symptoms such as a painful rash, swollen lymph nodes, and fever. Historically, monkeypox was confined to the heavily forested regions of West and Central Africa [1]. However, since May 2022, MPXV has spread globally with cases reported in countries that had no prior history of documented MPXV transmission. According to the World Health Organization (WHO), a total of 124,753 laboratory-confirmed cases were reported across 128 member states in all six WHO regions from 2022 to 2024 [2]. Rapid and early detection of MPXV is crucial for facilitating timely intervention and reduces the risk of disease transmission.
Traditional diagnostic methods for MPXV include virus culture, electron microscopy, and polymerase chain reaction (PCR). However, these assays are time-consuming and require well-equipped laboratories operated by skilled technicians. In contrast, isothermal amplification techniques, such as loop-mediated isothermal amplification (LAMP) and recombinase polymerase amplification (RPA), have been developed for use in resource-limited settings. While these methods offer the advantage of rapid amplification, they are prone to generating false-positive results [3]. Clustered regularly interspaced short palindromic repeats (CRISPR) technology, initially renowned for its role in gene editing, has emerged as a powerful molecular diagnostic tool for nucleic acid detection followed the discovery of the trans-cleavage activity of Cas nucleases, including Cas9, Cas12, and Cas13 [4]. With the guide RNA (gRNA), Cas nucleases bind to target sequences and cleave single-stranded DNA (ssDNA) or RNA (ssRNA) probes, resulting in fluorescence emission. Due to their sensitivity to mismatches and ability to detect single-base differences [5, 6], CRISPR/Cas-based detection systems exhibit exceptional specificity. To enhance sensitivity, isothermal amplification is often combined with CRISPR/Cas detection. Several platforms have been established based on this approach, such as SHERLOCK [5], DETECTR and HOLMES [7, 8]. These methods enable rapid and accurate detection of targets. However, they typically involve a two-step process, which requires opening the amplification tube and risks cross-contamination. To address this limitation, HOLMESv2 was developed by integrating a thermophilic endonuclease Cas12b with LAMP, enabling one-pot, one-step detection for practical applications [6].
Visual detection methods are vital for identification of target nucleic acids in resource-limited settings. In this study, through systematic screening of optimal primer sets and gRNAs coupled with optimization of critical reaction parameters including reaction temperature, buffer composition, Cas12b/gRNA, ssDNA reporter, and essential chemical additives, we proposed a streamlined, one-step visual detection assay for MPXV, which integrates the LAMP with CRISPR/Cas12b system within a single reaction vessel (Fig. 1). This assay enables the visualization of results with a vivid green fluorescence discernible to the naked eyes under blue light or ultraviolet (UV) illumination within 40 min using extracted viral nucleic acids. The simplicity and efficiency of this method render it suitable for deployment in regions where the MPXV is prevalent as long as there is a heat block or water bath and a portable blue light or UV flashlight.
Schematic illustration of the MPXV LAMP-CRISPR/Cas12b detection assay. A Workflow of the LAMP-CRISPR/Cas12b assay. B Working principle of LAMP-CRISPR/Cas12b assay. CRISPR-Cas12b specifically recognizes the targets and activates the trans-cleavage activity to generate fluorescent signals
According to the Technical Guidelines for Monkeypox Prevention and Control issued by the National Health Commission of China [9], samples were collected from suspected monkeypox cases meeting the following criteria: clinical presentation with fever, lymphadenopathy, and cutaneous/mucosal rash; plus at least one epidemiological risk factor within 21 days prior to symptom onset, including: (a) travel history to countries with reported monkeypox cases; (b) close contact with a confirmed monkeypox patient; or (c) exposure to blood, body fluids, or secretions from MPXV-infected animals (particularly rodents or non-human primates). A total of 113 clinical specimens comprising rash fluid and throat swabs were collected from suspected monkeypox cases during a one-year surveillance period (June 2023 to June 2024) in Jiangsu Province. All samples were immediately placed in viral transport medium (VTM) and stored at -80℃ in our laboratory freezer until use. The MPXV strains were obtained by inoculating Vero E6 cells with PCR-positive MPXV rash fluid swabs and cultured in a 37℃ 5%CO2 incubator. When typical cytopathic effects were observed and PCR confirmed positivity in the culture supernatant, the culture was passaged for further propagation. All virus isolation procedures were performed in a Biosafety Level 3 (BSL-3) laboratory. Viral nucleic acids were extracted from 200 µL VTM or virus culture using the Viral DNA/RNA Extraction Kit (T014, TIANLONG, China) with GeneRotex 96 (TIANLONG, China), and were stored at -80℃ until further use.
The LAMP primers were automatically designed using PrimerExplorer V5 (available at primerexplorer.jp/lampv5e/index.html) based on the F3L gene (Accession no: MT250197.1, 46192–46653). The gRNAs required for Cas12b-mediated trans-cleavage comprise a scaffold sequence(GUCUAGAGGACAGAAUUUUUCAACGGGUGUGCCAAUGGCCACUUUCCAGGUGGCAAAGCCCGUUGAGCUUCUCAAAUCUGAGAAGUGGCAC) at the 5′ end and a 20-nucleotide target-specific spacer sequence at the 3′ end with a protospacer adjacent motif (PAM) sites of 5′ -TTN-3′ or 5′ -TAC-3′ as previously described [6]. All primers and spacer sequences of gRNAs were submitted for a BLAST search to ensure alignment was exclusive to the MPXV (https://www.ncbi.nlm.nih.gov/tools/primer-blast/). The detailed sequences of the primers and gRNAs designed in this study are provided in Supplementary Table S1&S2, and were synthesized by Sangon Biotech Co., Ltd. (China).
The gRNAs used in this study were prepared by in vitro transcription with synthesized DNA template, which was the reverse complementary sequence of gRNA, with the reverse complementary sequence of the T7 promoter (5’-CCCTATAGTGAGTCGTATTA-3’) at the 3’ end. The reaction mixture comprising 1 µL DNA template (500 ng/µL), 1 µL T7 primer (5’-TAATACGACTCACTATAGGG-3’) (40 µM), 5 µL T7 RNA Polymerase Buffer, and 30.25 µL of nucleases-free water was reacted at 95℃ 30s, 25℃ 2 min, then, 12.75 µL of master mix containing 2.5 µL T7RNA Polymerase (50 U/µL) (2540 A, TaKaRa), 5 µL DTT (50 mM), 4 µL NTPs (25 mM), 1.25 µL Recombinant Rnase Inhibitor (40 U/µL) was added immediately and transcribed at 42℃ overnight. The transcribed gRNAs were treated with 20 U Recombinant DNase I (2270 A, TaKaRa) at 37℃ 1.5 h to remove the DNA template, followed by purification via Spin Column RNA Cleanup & Concentration Kit (B518688, Sangon, China). The concentration was quantified by the Qubit Flex Fluorometer via the Qubit RNA Broad Range Assay Kits (Q10210, Thermo, USA), and stored at -80℃ until use.
The 10× LAMP Primer Mix containing 2 µM each of F3/B3, 16 µM each of FIP/BIP, 4 µM each of LF/LB was prepared in nucleases-free water and store at − 20℃ until use. The LAMP was performed at 65℃ following the manufacture’s instruction by using 25 µL of reaction mixture containing 12.5 µL WarmStart LAMP 2× Master Mix (E1700S, New England Biolabs, USA), 2.5 µL 10 ×LAMP Primer Mix, 1 µL template and 9 µL nucleases-free water. The amplicons were monitored by the Loopamp LA-500 (Eiken, Japan) real-time turbidity instrument.
The one-pot two-step CRISPR/Cas12b detection was employed to screen for optimal gRNAs. Briefly, 20 µL LAMP reaction mixture including 1 µL template DNA was added at the bottom of tube. 5 µL Cas12b cleavage system containing 5 µM AapCas12b (32118, Tolo Biotechnology, China), 5 µM gRNA, 6.25 µM ssDNA reporter (5’-6-FAM-TTTTT-BHQ1-3’), and 2.5 µL 10×Holmes buffer 1 was added in the cap. Following a 30-minute LAMP amplification, Cas12b cleavage system was mixed with the LAMP products through brief centrifugation. Fluorescence signals were collected by CFX96 Deep Well real-time system with C1000 Thermal Cycler module (Bio-rad, United States) at 60℃ every 1 min. Upon completion of the reaction, the tubes were examined under a blue light transilluminator (OSE-470 L, TIANGEN BIOTECH, China), and images were captured using a smartphone. Additionally, the tubes were photographed under UV light using an Azure Biosystems c280 imaging system (United States).
To achieve a one-step, one-pot integration of LAMP amplification and CRISPR/Cas12b detection, both the LAMP reaction mixture and the CRISPR system were added into the bottom of the same PCR tube. The key parameters, including the ratio of reaction buffers, reaction temperature, concentrations of Cas12b/gRNA, and ssDNA reporter, as well as the effects of chemical additives including glycine and taurine (Aladdin, China) were systematically optimized. Fluorescence signals were monitored using the Bio-rad CFX96 Deep Well real-time PCR system.
The MPXV DNA was quantified using digital PCR following the protocol provided with the QIAcuity Probe PCR Kit (250101, QIAGEN, Germany). To assess the detection sensitivity, MPXV DNA was serially diluted 10-fold, covering a range from 104 to 100 copies. In parallel, the limit of detection (LOD) for both the fluorescent and visual CRISPR/Cas12b-LAMP assays were determined by testing MPXV DNA at a narrower range of concentrations (25, 10, 5, and 1 copies) in eight replicates, with nucleases-free water serving as the negative control. The 95% probability detection limit was calculated using probit regression analysis in SPSS.
To evaluate the specificity of the assay, herpes simplex virus (HSV), Epstein-Barr virus (EBV), measles virus (MV), and enteroviruses CVA16 and EV-A71, which can cause rashes similar to those of monkeypox, were tested with LAMP-CRISPR/Cas12b. MPXV was used as the positive control to ensure the assay’s ability to distinguish MPXV from these potentially confounding pathogens.
A total of 113 clinical samples were analyzed using both LAMP-CRISPR/Cas12b and qPCR for comparative evaluation. The qPCR was conducted in accordance with the Technical Guidelines for Monkeypox Prevention and Control issued by the National Health Commission of China [9]. The reaction mixture consisted of 10 µL Premix Ex Taq (Probe qPCR) (2×) (Takara), 0.4 µL each of forward and reverse primers and probe (10 µM), 4 µL of template, and 4.8 µL of nucleases-free water. The thermal cycling conditions were as follows: initial denaturation at 95℃ for 30s, followed by 40 cycles of 95℃ for 5s and 60℃ for 30s. Fluorescence was collected using the Bio-rad CFX96 Deep Well real-time PCR system.
The graphs were drawn by the GraphPad Prism 5, data statistics were performed using IBM SPSS Statistics 23.0 Standard Network Version.
The F3L gene was selected as a target due to its relatively high conservation among MPXV, five LAMP primer sets (PS1-PS5) were designed targeting the F3L (Table S1). Primer set 5 (PS5) demonstrated the shortest take-off time for MPXV DNA and no amplification curve for the negative control within 1 h (Fig. 2A). Based on the amplification products of Primer set 5, five Cas12b gRNAs were designed (Table S2). As shown in Fig. 2B, both gRNA4 and gRNA5 induced good trans-cleavage activity of cas12b, and gRNA5 with higher fluorescence signals was selected for subsequent experiments. The location of primer set 5 and gRNA 5 binding sites was shown in Fig. 2C.
LAMP primer and gRNA screening. A The LAMP primer sets screening. B The gRNAs screening in the two-step LAMP-CRISPR/Cas12b assay. NC, negative control. C LAMP primer set 5 and gRNA 5 design regions used in the study. A part of the F3L sequence (bp 180 to 420) is shown. The spacer sequence of gRNA 5 was marked in grey area, right-pointing arrows and left-pointing arrows indicate sense and complementary strand of primer set 5
Due to the operational complexity of the two-step LAMP-CRISPR/Cas12b, we aimed to optimize the detection protocol into one-step. As shown in Fig. 3A, a combination of 0.5×LAMP master mix and 0.5×CRISPR buffer (10×holmes buffer 1) yielded the best detection performance. The reaction temperature was tested between 58℃ and 65℃. Results indicated that 60℃ generated the fastest positive results, whereas no amplification signals were detected at 62℃ and 65℃ with 100 copies of MPXV DNA as template (Fig. 3B). For Cas12b/gRNA concentrations, dilutions from 0.05 µM to 0.5 µM were tested. The shortest threshold time was observed at 0.2 µM, with no obvious nonspecific fluorescence in the negative control (Fig. 3C). Regarding ssDNA probes, although higher concentrations enhanced fluorescence signals, they also increased nonspecific signals in negative controls, raising the risk of false positives. Therefore, 0.25 µM was chosen as the optimal concentration for detection (Fig. 3D).
The optimization of various parameters in the one-step, one-pot LAMP-CRISPR/Cas12b assay, including the proportions of reaction components (A), reaction temperatures (B), concentrations of AapCas12b/gRNA complexes (C), and ssDNA reporters (D) with 100 copies of MPXV DNA as template. NC, negative control. LAMP and CRISPR here are WarmStart LAMP 2×Master Mix and 10×homelsbuffer 1, respectively
Additionally, we explored the effects of chemical additives on the efficiency of LAMP-CRISPR/Cas12b assay. Results revealed that 0.25 M glycine or 0.1 M taurine significantly boosted fluorescence signals and reduced threshold time, while the combined use of 0.25 M glycine and 0.1 M taurine did not yield better results (Fig. 4).
Fluorescence signals over time under varying final concentrations of glycine (A), taurine (B), and their combination (C) in the one-step LAMP-CRISPR/Cas12b assay. NC, negative control
Finally, the one-step, one-pot LAMP-CRISPR/Cas12b assay for MPXV diagnosis we established was performed at 60℃ with 20 µL reaction mix containing 5 µL WarmStart LAMP 2×Master Mix, 1 µL 10×homelsbuffer 1, 2 µL 10×LAMP Primer Mix, 0.2 µM gRNA, 0.2 µM AapCas12b, 0.25 µM ssDNA reporter, 0.25 M glycine, 1 µL template and nucleases-free water.
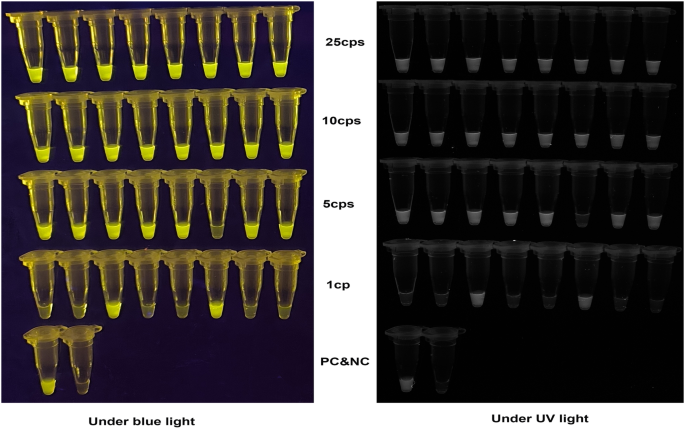
The result showed that the threshold time (tt) of the assay gradually increased with decreasing DNA concentration (104-100 copies) (Fig. 5A), and was inversely proportional to the log values of the target concentration (copies), demonstrating good linearity for MPXV quantification (R2 = 0.932) (Figure S1). The amplification curves appeared within 20 min even at 10 copies, and bright green fluorescence was clearly visible to the naked eyes under both blue light and UV light illumination after 40 min reaction (Fig. 5C). At 25 copies of MPXV DNA, both the fluorescent and visual LAMP-CRISPR/Cas12b assays achieved 100% positive detection (8/8 reactions), for 10, 5 copies and 1 copy per reaction, the positive detection rates were 100% (8/8), 62.5% (7/8), 25% (2/8), respectively (Fig. 6). Probit regression analysis determined that the LOD of the one-step LAMP-CRISPR/Cas12b assay was 6.5 copies per reaction.
Sensitivity of the one-step LAMP-CRISPR/Cas12b assay. Real-time (A) and endpoint fluorescence verified by fluorescence detection (B) as well as visual detection (C) with 10-fold gradually diluted MPXV DNA (104-100 copies) as templates for 40 min incubation. cps, copies; NC, negative control
Limit of detection of the visual one-step LAMP-CRISPR/Cas12b assay. Limit of detection was assessed using decreasing input (25, 10, 5, 1 copies) of MPXV DNA with eight replicates. cps, copies; PC, positive control with 100 copies of MPXV DNA. NC, negative control
No amplification signal was detected in the nucleic acids of HSV, EBV, MV, CVA16, EV-A71 tested, while MPXV demonstrated good amplification curve (Fig. 7), which proves the high specificity of our assay.
Specificity of the one-step LAMP-CRISPR/Cas12b assay. Real-time (A) and endpoint fluorescence verified by fluorescence detection (B) as well as visual detection (C) with viral templates of HSV, EBV, MV, CVA16, EV-A71, and MPXV for 40 min incubation. cps, copies; NC, negative control
To evaluate the clinical applicability of our method for MPXV detection, 113 clinical samples were analyzed using both LAMP-CRISPR/Cas12b and qPCR for comparative evaluation. The LAMP-CRISPR/Cas12b exhibited a sensitivity of 100% (71/71) and a specificity of 100% (42/42) compared to qPCR (Table 1). Threshold times of all positive clinical samples were consistently below 20 min (Fig. 8), and can be visualized by naked eyes under blue light or UV light (Figure S3).
Clinical validation of the one-step LAMP-CRISPR/Cas12b assay. Scatter plot of the tt (time to threshold) values of one-step LAMP-CRISPR/Cas12b assay and the Ct values of the real-time qPCR assay on clinical samples. Each dot indicates one sample
The apparent increase in human monkeypox cases across a wide geographic area and the potential for further spread have heightened concerns regarding this emerging disease. Early diagnosis is crucial to prevent the rapid global dissemination. Nucleic acid testing such as PCR represents the most sensitive and specific method for the early detection of pathogens, but unsuitable for rapid point-of-care testing due to its reliance on sophisticated instruments and trained personnel. In this study, we developed a novel on-site rapid and simple assay for MPXV detection by integrating the LAMP with CRISPR/Cas12b system in one-pot. Through optimization of critical reaction parameters, the assay showed excellent concordance with qPCR, but the LAMP-CRISPR/Cas12b only took 40 min.
In one-pot testing, Cas12b can inhibit LAMP amplification by cleaving both the MPXV dsDNA and its amplicons [10]. Therefore, the reaction buffer is a key factor in the establishment of the one-pot assay for combining LAMP amplification with CRISPR/Cas12b detection. We determined that a combination of 0.5×LAMP mix and 0.5×CRISPR buffer yielded the best detection performance. Due to the inherent limitation of the CRISPR system, which can only detect nucleic acids at the fM level (~ 602 copies/µL) without prior amplification [11]. The fluorescence curve of 10-100cps MPXV in our study indicated that LAMP amplification predominated during the initial phase. When the nucleic acid amplification products reached a certain concentration, the cleavage activity of Cas12b was no longer effectively inhibited, yielding a detectable fluorescence signals. Additionally, our results revealed that the addition of glycine or taurine significantly boosted fluorescence signals and reduced threshold time, consistent with the previous report [12]. Previous study revealed that glycine effectively boosted the amplification rate of LAMP for HBV detection in the one-pot assay [13], which indicated that the amino acid additives might function as protein stabilizer to maintain the activity of the Bst DNA polymerase of the LAMP. Furthermore, the purity of in vitro transcribed gRNA is crucial, as residual DNA templates can lead to elevated fluorescence in negative controls. To mitigate this issue, commercially synthesized gRNAs may be a more reliable choice.
We reviewed studies reporting point-of-care visual molecular detection methods for MPXV during the recent three years (Table S3), most of these methods were based on LAMP-LFB [14] or colorimetric LAMP which detected by-products of LAMP amplification, such as H+-induced pH changes (e.g., from pink to yellow) [15,16,17], or pyrophosphate-mediated sequestration of metal ions in complex with dyes (e.g., 5-Br-PAPS or calcein, resulting in color changes from red to yellow or orange to green) [18, 19]. However, these approaches often face challenges with false positives due to interactions among 4 to 6 primers of LAMP or self-amplifying hairpins. Additionally, colorimetric methods tend to have high background interference for naked-eye observation, as the color changes typically occur between adjacent hues. Several CRISPR-based methods for MPXV detection have been developed to improve specificity [20,21,22,23,24], However, these methods often require separate reaction steps, increasing the risk of cross-contamination and operational complexity. In contrast, our proposed method integrates LAMP with CRISPR in a one-pot, one-step reaction, eliminating the risk of cross-contamination. Leveraging the high amplicon yield of LAMP (10 to 100 times more sensitive than conventional PCR) [25], our method achieves a limit of detection (LOD) of 6.5 copies per reaction, exhibiting superior sensitivity compared to the vast majority of existing isothermal visual detection methods of MPXV. Furthermore, the Cas12b-mediated trans-cleavage of fluorescence-quenched reporters, producing a stable bright green fluorescence in positive samples that remains distinguishable for several days, which is more visually discernible than colorimetric methods. In terms of clinical applicability, our method offers a significant advantage over previously reported methods, as it has been validated using 113 clinical samples rather than simulated ones. This underscores its potential for real-world diagnostic applications. Furthermore, the good linearity observed in the standard nucleic acid curve demonstrates the feasibility of semi-quantitative analysis for MPXV samples.
A limitation of this method is that only MPXV Clade II clinical samples were used for validation. To further assess the robustness of our method, clinical samples from MPXV Clade I should be included if available in the future. Additionally, the nucleic acids used in this study were extracted using an automated nucleic acid extraction system. However, the potential impact of various nucleic acid extraction approaches including automated extraction, manual extraction methods (e.g., phenol-chloroform extraction or silica column centrifugation), and extraction-free processing techniques (such as heat treatment of unextracted diagnostic samples to obliterate nucleases, HUDSON, etc.) on detection performance remains to be systematically evaluated. Further investigation incorporating extraction-free methodologies is warranted to enhance the assay’s adaptability for point-of-care applications.
Given that the essential reaction components can be pre-prepared through lyophilization, necessary equipments (a heat block or water bath and a blue or UV flashlight) and regents can be conveniently packed into a small portable case. This enables our method to function as an on-site detection system for MPXV. Consequently, the proposed method holds significant potential for point-of-care testing outside traditional clinical diagnostic laboratories, such as at airports or outbreak sites, providing a practical and efficient solution for rapid MPXV detection in resource-limited settings.
In conclusion, we developed a novel on-site rapid and simple assay for MPXV detection by integrating the LAMP with CRISPR/Cas12b system in one-pot, which showed excellent concordance with qPCR. The simplicity and efficiency of this method renders it suitable for deployment in regions where the MPXV is prevalent as long as there is a heat block or water bath and a portable blue light or UV flashlight.
No datasets were generated or analysed during the current study.
- MPXV:
-
Monkeypox virus
- LAMP:
-
Loop-mediated isothermal amplification
- qPCR:
-
Quantitative PCR
- LOD:
-
Limit of detection
- WHO:
-
World Health Organization
- RPA:
-
Recombinase polymerase amplification
- CRISPR:
-
Clustered regularly interspaced short palindromic repeats
- gRNA:
-
Guide RNA
- UV:
-
Ultraviolet
- BSL-3:
-
Biosafety Level 3
- PAM:
-
Protospacer adjacent motif
Not applicable.
This study was supported by the National Key R&D Program of China (2023YFC2605100, 2023YFC2605104) and the natural science foundation of Jiangsu Province (BK20231374, BK20221413).
This study has been approved by the ethical Review Committee of Jiangsu Provincial Center for Disease Control and Prevention. Informed consent was provided by all participants. All the experiments were carried out in accordance with the Declaration of Helsinki.
All authors consent for publication.
The authors declare no competing interests.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
Guo, J., Shan, Y., Hu, G. et al. Rapid visual detection of Monkeypox virus by one-step LAMP-CRISPR/Cas12b assay. Virol J 22, 151 (2025). https://doi.org/10.1186/s12985-025-02780-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12985-025-02780-0